Posted on April 2, 2016
By John Jones
Journal articles
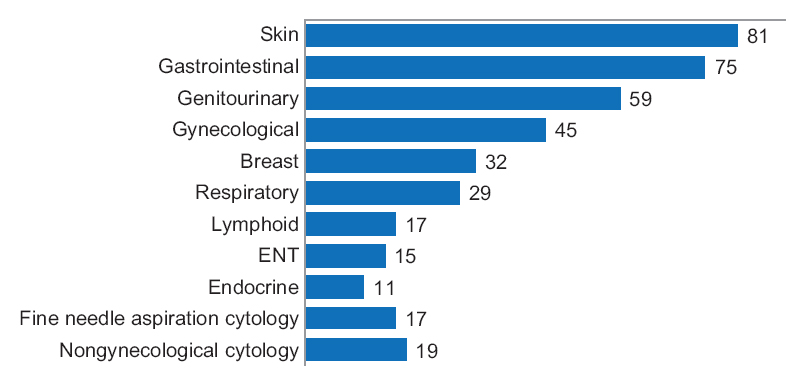
Is digital diagnostic determination of pathology images quicker than microscopic? It depends on who you ask. Vodovnik published results of his comparative study in early 2016, comparing digital and microscopic diagnostic times using tools such as digital pathology workstations and laboratory information management systems. His conclusion: "A shorter diagnostic time in digital pathology comparing with traditional microscopy can be achieved in the routine diagnostic setting with adequate and stable network speeds, fully integrated LIMS and double displays as default parameters, in addition to better ergonomics, larger viewing field, and absence of physical slide handling, with effects on the both diagnostic and nondiagnostic time."
Posted on March 26, 2016
By John Jones
Journal articles
In 2010, Philip Wenig and Juergen Odermatt published their experiences on developing OpenChrom, an open-source chromatography data management system (CDMS) for analyzing mass spectrometric chromatographic data. The team built the software due to a perceived "lack of software systems that are capable to enhance nominal mass spectral data files, that are flexible, extensible and that offer an easy to use graphical user interface." The group concluded that "OpenChrom will be hopefully extended by contributing developers, scientists and companies in the future." As of 2016, development on OpenChrom does indeed continue, with a 1.1.0 preview release available released in February.
Posted on March 23, 2016
By John Jones
Journal articles

This 2012 paper, published in
Journal of Pathology Informatics, provides the perspective of Yale University School of Medicine's Pathology Informatics Unit in regards to their experience with custom software development. Despite perceptions concerning the pitfalls of laboratories developing their own software, Sinard
et al. conclude that "[m]any of the risks associated with custom development can be mitigated by a well-structured development process, use of open-source tools, and embracing an agile development philosophy."
Posted on March 15, 2016
By John Jones
Journal articles
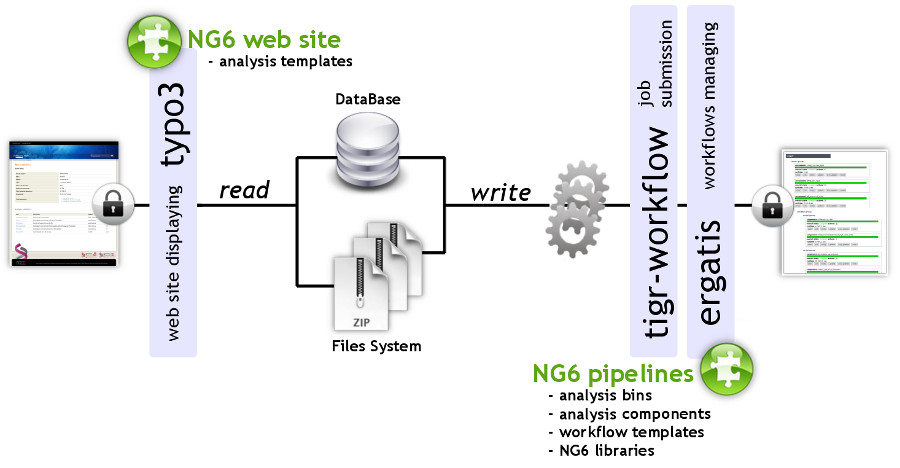
Published in 2012, this paper on the NG6 next generation sequencing (NGS) platform describes the software's development process at GenoToul, a French shared life sciences research facility. Mariette
et al. found that NGS software Galaxy was promising but not sufficient as it only "aims at simplifying data processing for researchers," whereas the group needed a LIMS that was also good at "gathering specialized pipelines and website" data. NG6 was developed to meet that need.
Posted on March 14, 2016
By John Jones
Journal articles

Hernández-de-Diego et al. from the Centro de Investigación Príncipe Felipe and the Karolinska Institute needed a softwar that " [i]n contrast to other solutions that put the focus on management of thousands of samples for core sequencing facilities ... [could handle] the annotation of experiments designed and run at individual research laboratories." Additionally, genomics solutions such as BASE had annotation limitations in regards to microarray experiment data. The group decided to develop their own system called the STATegra experiment management system (EMS), which they concluded "provides an integrated system for annotation of complex high-throughput omics experiments at functional genomics research laboratories."
Posted on March 3, 2016
By John Jones
Journal articles

In this 2012 journal article from
ZooKeys, Blagoderov and his colleagues from the Natural History Museum in London describe the process they went through to improve the mass-digitization of their numerous collections. Noting problems with cost, "as well as the inherent fragmentation in collection based biodiversity informatics," the researchers created a "wall-to-wall" process using the SatScan tray scanning system. The group suggested that such a system could, when implemented well, "open up collections to the world, facilitating their use, and help create a global collection index that can be used to set priorities for further digitization."
Posted on March 3, 2016
By John Jones
Journal articles
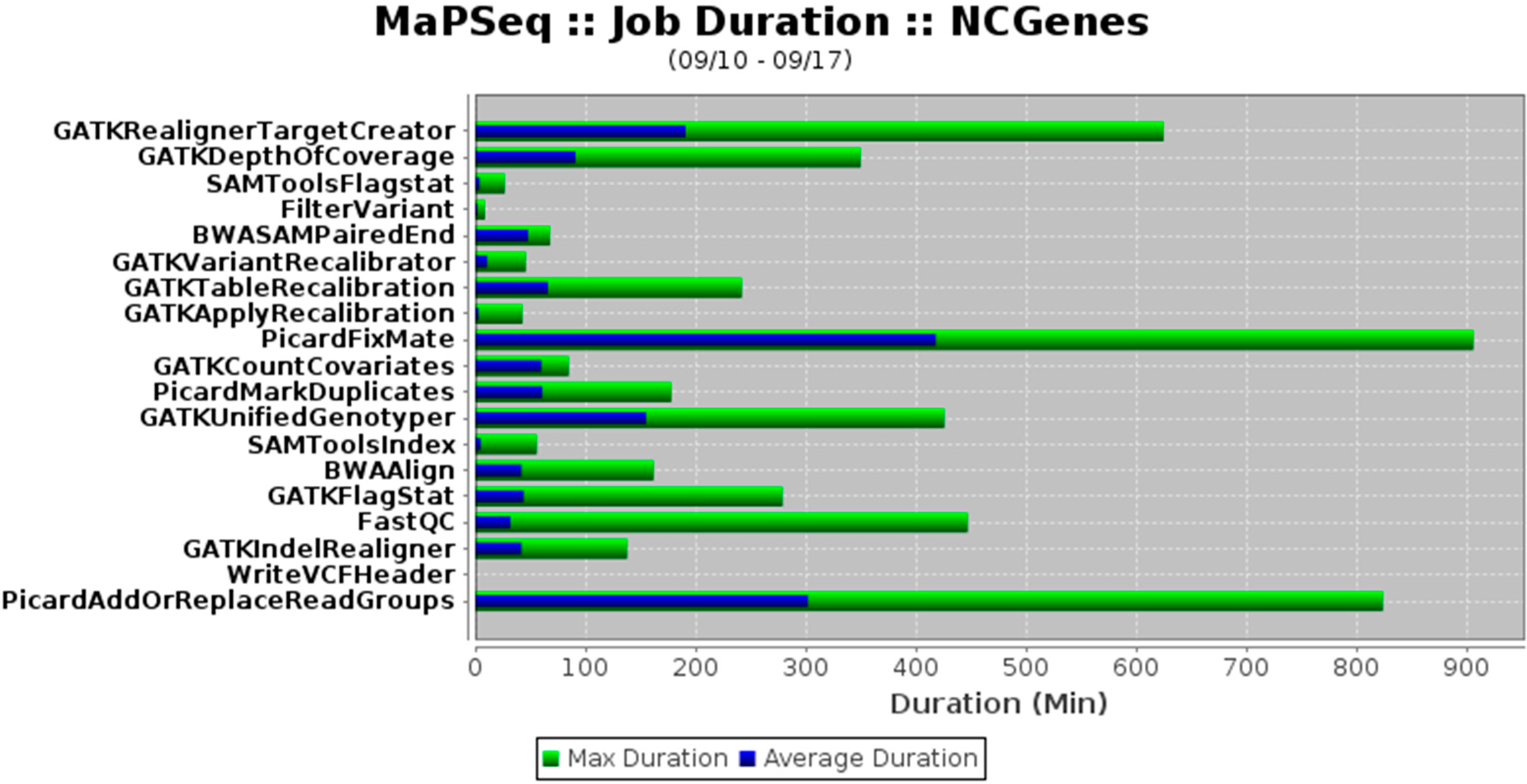
Reilly et al. at the University of North Carolina needed a specific solution for their genomics research, a solution that would "address the organizational challenges of computation-intensive biomedical research within a decentralized academic institution." After reviewing numerous options, the team decided to create their own system, MaPSeq, an application built on service-oriented architecture. In this paper summing up their development and implementation process, the group concludes that while the software is useful for massively parallel sequencing, "the general architecture and approach can be adapted for other complex or computationally-intense workflows."
Posted on February 15, 2016
By John Jones
Journal articles

In this brief article published in
Frontiers in Environmental Science in 2014, environmental researcher Alexander Kokhanovsky takes a look at the state of environmental informatics past and present while also speculating about the future challenges. He ends by emphasizing the need for improved "estimation of aerosol load using space-borne instrumentation" to improve environmental research, including improved algorithms and visualization tools.
Posted on February 12, 2016
By John Jones
Journal articles
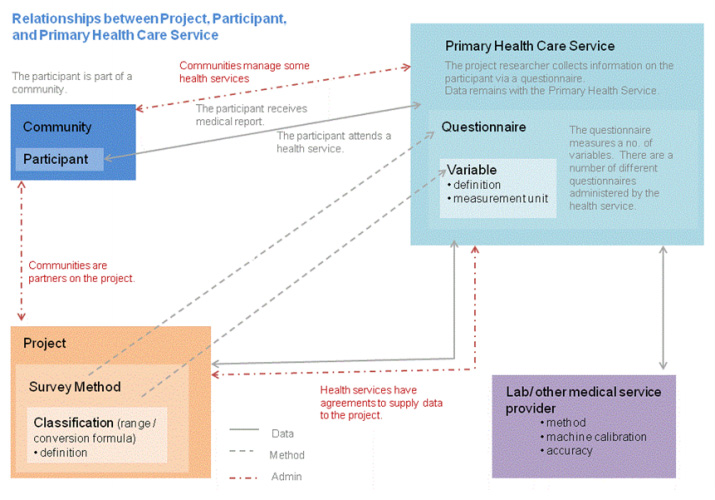
van Gaans
et al. of the University of South Australia and James Cook University needed a system that provides data management functionality for the specific needs of a public health research organization, including data sharing and reuse. They needed a system that would "ensure that: data that are unmanaged be managed, data that are disconnected be connected, data that are invisible be findable, [and] data that are single use be reusable, within a structured collection." In this journal article, the group documents the development of their answer: the Public Health Data Management System (PHRDMS).
Posted on January 28, 2016
By John Jones
Journal articles
Forensic science researcher Bruce Levy of the University of Illinois at Chicago presents his ideas about how forensic pathologists and clinical informaticians can address the historical and current shortcomings of putting sudden and violent death data to better use. Levy argues that improved collaborations and data standards "will enable forensic pathology to maximize its effectiveness by providing timely and actionable information to public health and public safety agencies."
Posted on January 21, 2016
By John Jones
Journal articles
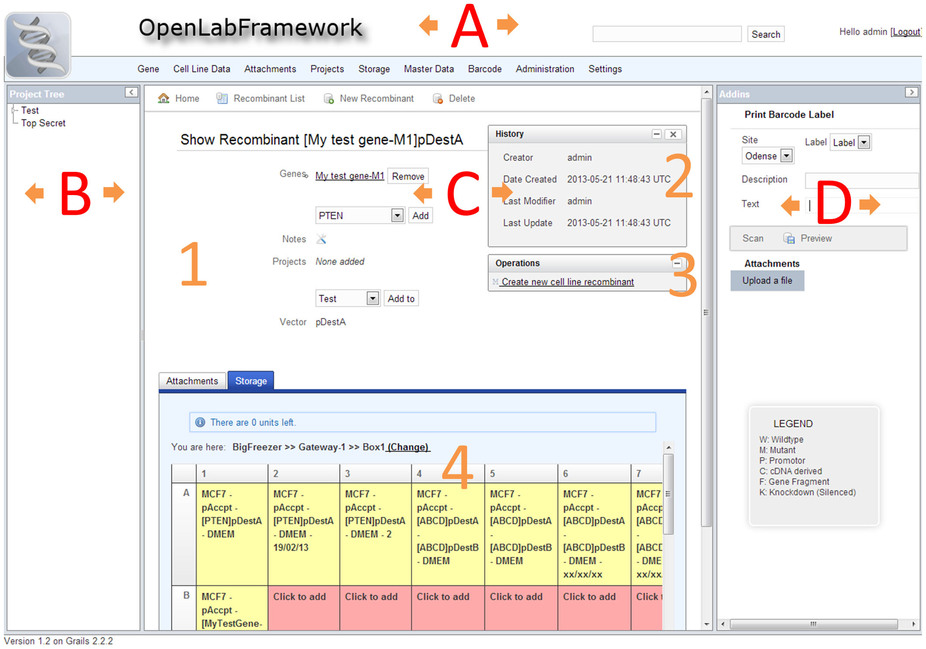
Nanomedicine researchers at the University of Southern Denmark required a "dedicated LIMS for the management of large vector construct and cell line libraries." With no other open-source options available, the team developed their own, OpenLabFramework. Documenting their process, List
et al. conclude "OLF can be deployed using different database management systems either locally, to a server, or to the cloud," and "[t]he incorporation of modern technologies, such as mobile devices and printing of barcode labels may increase productivity even further."
Posted on January 12, 2016
By John Jones
Journal articles
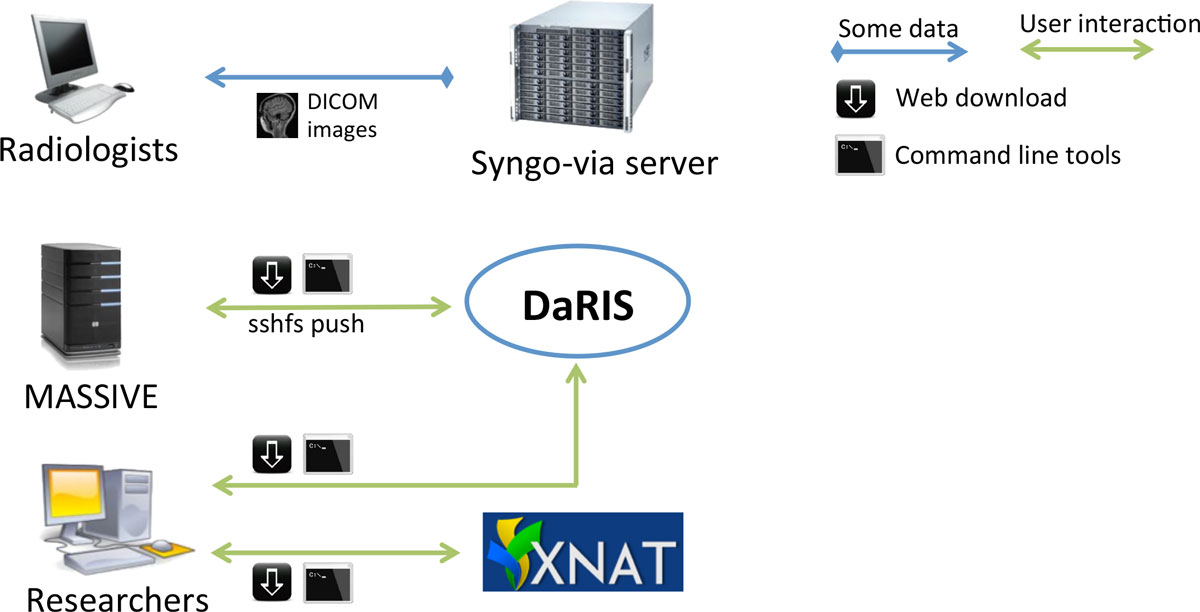
In this research paper, Nguyen et al. of Monash University in Australia "describes the design, implementation and operation of a multi-modality research imaging data management system that manages imaging data obtained from biomedical imaging scanners" at their facility. Faced with limitations of existing image management software and frameworks, the group custom built a system "based on DaRIS and XNAT has been designed and implemented to enable researchers to acquire, manage and analyse large, longitudinal biomedical imaging datasets."
Posted on January 11, 2016
By John Jones
Journal articles
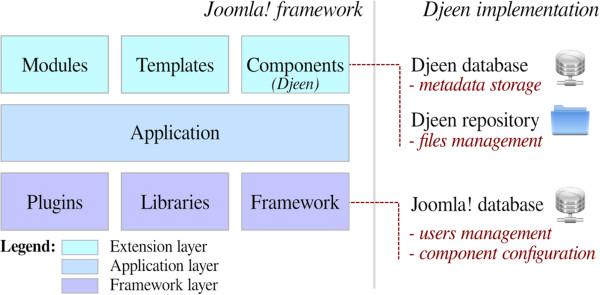
Stahl et al. and Universités de Montpellier needed a system that could "streamline [biological] data storage and annotation collaboratively." Not finding a system to their liking, the group developed a Joomla!-based LIMS called Djeen and published their research on the development process in 2013. The group concludes: "Djeen allows managing project associated with heterogeneous data types while enforcing annotation integrity and minimum information. Projects are managed within a hierarchy and user permissions are finely-grained for each project, user and group."
Posted on December 28, 2015
By John Jones
Journal articles
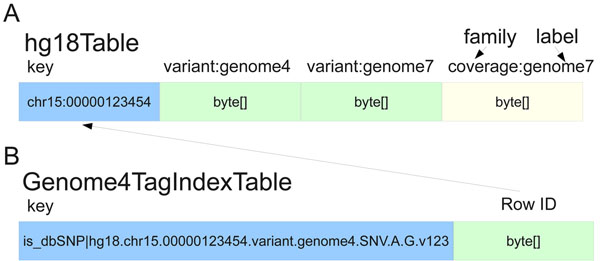
In 2010, O'Connor et al. published a paper on their experience developing and implementing a cloud-based query engine that can support thousands of genome datasets. Still actively developed as of 2015, the SeqWare Query Engine "can load and query variants (SNVs, indels, translocations, etc) with a rich level of annotations including coverage and functional consequences." The research team concluded in their paper that the software was then "capable of supporting large scale genome sequencing research projects involving hundreds to thousands of genomes as well as future large scale clinical deployments utilizing advanced sequencer technology that will soon involve tens to hundreds of thousands of genomes."
Posted on December 23, 2015
By John Jones
Journal articles

In 2015, Singh et al. published their notes and reflections on developing SaDA, designed "for storing, retrieving and analyzing data originated from microorganism monitoring experiments." The group developed the software after discovering a lack of free, open-source software for microarray data management and analysis. The group concluded that "the platform has the potential to become an appropriate tool for a wide range of users focused not only in water based environmental research but also in other studies aimed at exploring and analyzing complex ecological habitats."
Posted on December 23, 2015
By John Jones
Journal articles
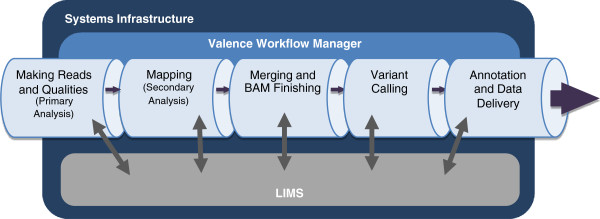
Seeking to overcome some of the challenges of massively parallel DNA sequencing, Reid et al. developed the Mercury analysis pipeline and deployed it on Amazon Web Services. Publishing their results in 2014 in
BMC Bioinformatics, the group demonstrated the success of "a powerful combination of a robust and fully validated software pipeline and a scalable computational resource that, to date, we have applied to more than 10,000 whole genome and whole exome samples."
Posted on December 17, 2015
By John Jones
Journal articles

Appearing in the magazine
Open Source Business Resource (today called
Technology Innovation Management Review) in 2011, this non-journal article by Sandro Groganz describes how "[o]pen source vendors can benefit from business ecosystems that form around their products," using open-source OXID eShop and its vendor OXID eSales as a representative example. Groganz concludes that "an open source community is not a state but a process," one that "creates a foundation for long-term growth and sustainability" when appropriately embraced.
Posted on December 16, 2015
By John Jones
Journal articles
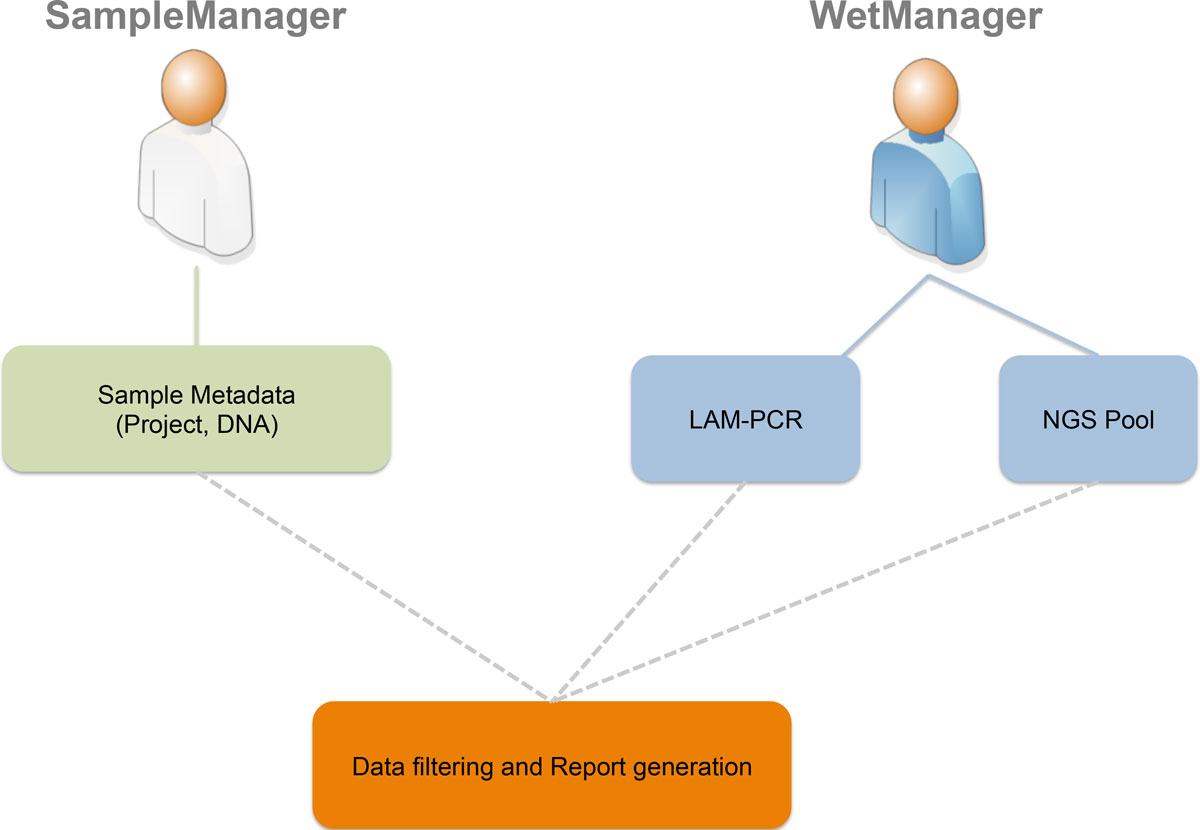
The San Raffaele Telethon Institute for Gene Therapy wanted a new LIMS to manage the data coming from their PCR techniques and next generation sequencing (NGS) methods. Not finding something suitable, the group developed its own LIMS, adLIMS. This 2015 research paper by Calabria et al., published in
BMC Bioinformatics, covers the academic aspects of the information collection and development process.
Posted on December 9, 2015
By John Jones
Journal articles
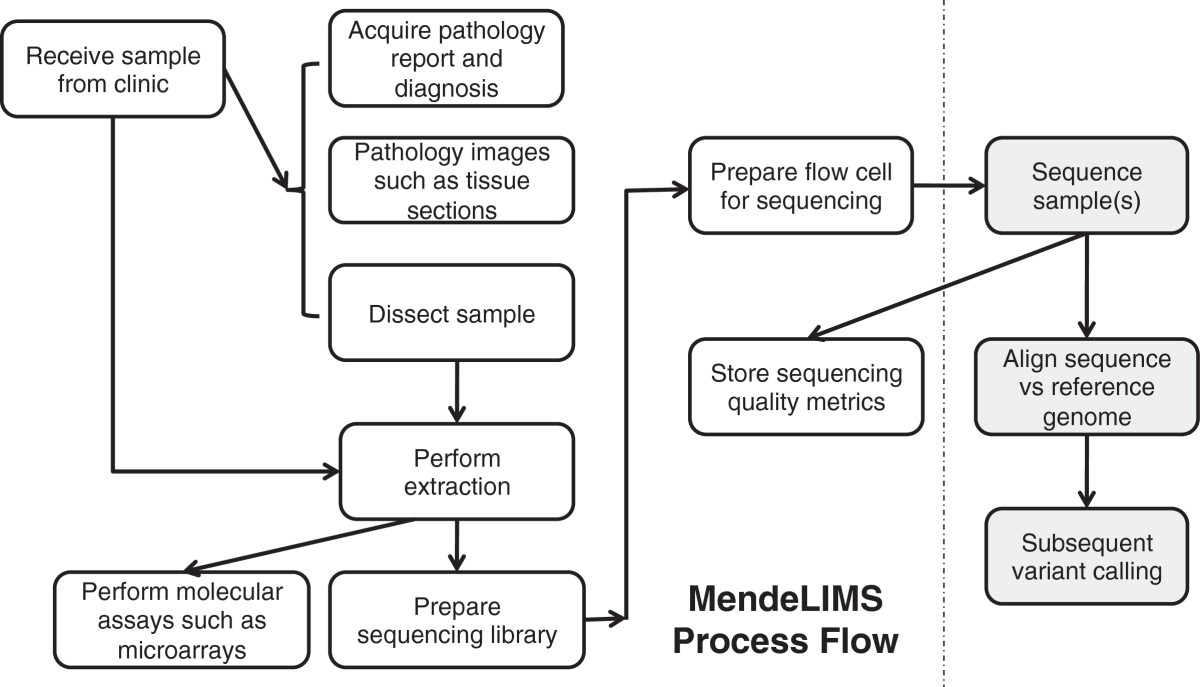
This 2014 research published in
BMC Bioinformatics sees Grimes and Ji presenting their "highly configurable and extensible" web-based laboratory information management system (LIMS) for next generation DNA sequencing, MendeLIMS. The group concludes that the software is "an invaluable tool for the management of our clinical sequencing studies," primarily due to its ability to reduce sample tracking errors and give "a comprehensive view of data being sequenced."
Posted on December 2, 2015
By John Jones
Journal articles
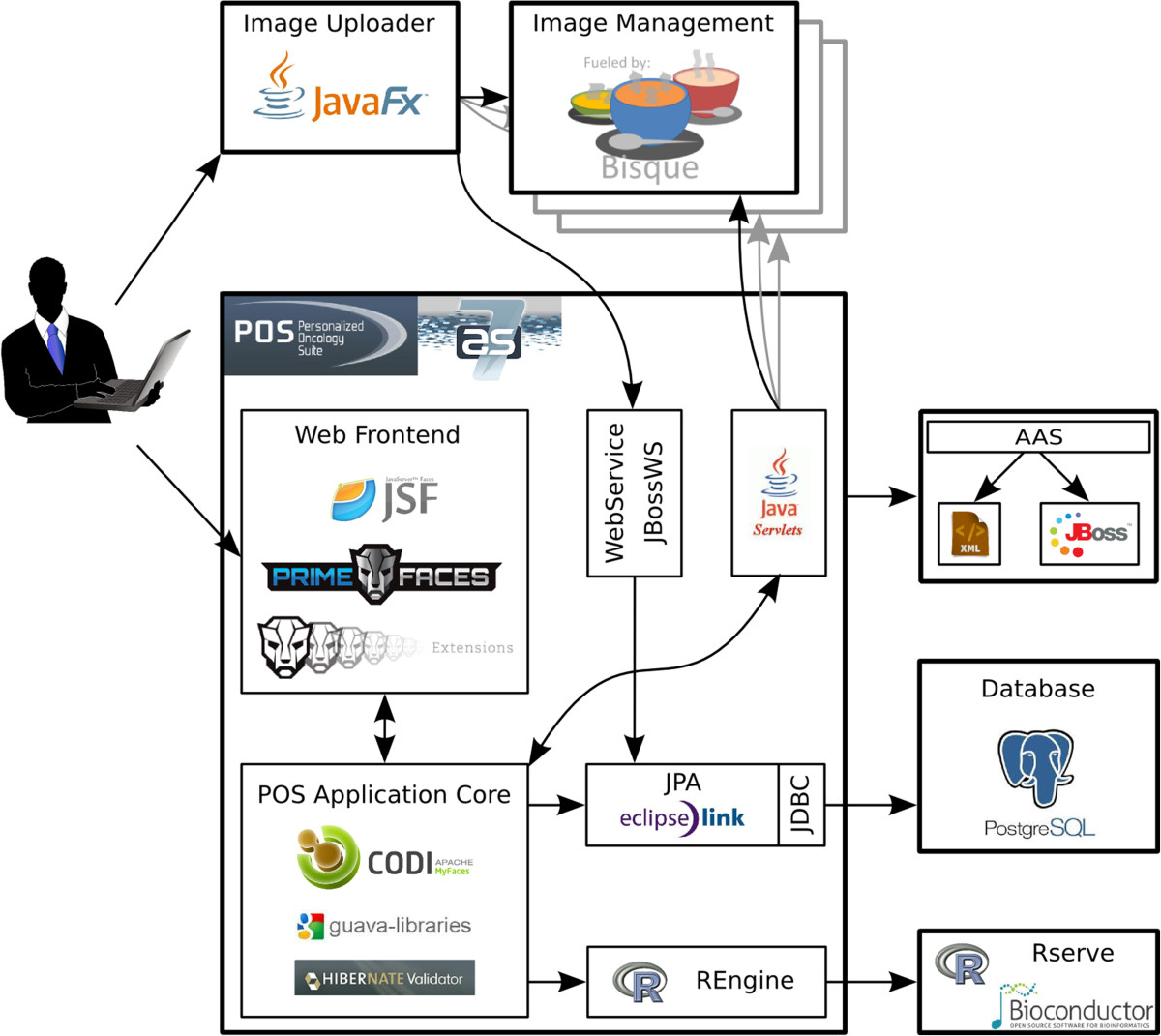
In this 2014 article published in
BMC Bioinformatics, Dander et al., not content with the disparity among existing cancer treatment and bioinformatics platforms, discussed the results of creating Personalized Oncology Suite (POS). The web-based, scalable software system "integrates clinical data, NGS data and whole-slide bioimages from tissue sections" and, as the team concludes, "can be used not only in the context of cancer immunology but also in other studies in which NGS data and images of tissue sections are generated."
 Is digital diagnostic determination of pathology images quicker than microscopic? It depends on who you ask. Vodovnik published results of his comparative study in early 2016, comparing digital and microscopic diagnostic times using tools such as digital pathology workstations and laboratory information management systems. His conclusion: "A shorter diagnostic time in digital pathology comparing with traditional microscopy can be achieved in the routine diagnostic setting with adequate and stable network speeds, fully integrated LIMS and double displays as default parameters, in addition to better ergonomics, larger viewing field, and absence of physical slide handling, with effects on the both diagnostic and nondiagnostic time."
Is digital diagnostic determination of pathology images quicker than microscopic? It depends on who you ask. Vodovnik published results of his comparative study in early 2016, comparing digital and microscopic diagnostic times using tools such as digital pathology workstations and laboratory information management systems. His conclusion: "A shorter diagnostic time in digital pathology comparing with traditional microscopy can be achieved in the routine diagnostic setting with adequate and stable network speeds, fully integrated LIMS and double displays as default parameters, in addition to better ergonomics, larger viewing field, and absence of physical slide handling, with effects on the both diagnostic and nondiagnostic time."
 In 2010, Philip Wenig and Juergen Odermatt published their experiences on developing OpenChrom, an open-source chromatography data management system (CDMS) for analyzing mass spectrometric chromatographic data. The team built the software due to a perceived "lack of software systems that are capable to enhance nominal mass spectral data files, that are flexible, extensible and that offer an easy to use graphical user interface." The group concluded that "OpenChrom will be hopefully extended by contributing developers, scientists and companies in the future." As of 2016, development on OpenChrom does indeed continue, with a 1.1.0 preview release available released in February.
In 2010, Philip Wenig and Juergen Odermatt published their experiences on developing OpenChrom, an open-source chromatography data management system (CDMS) for analyzing mass spectrometric chromatographic data. The team built the software due to a perceived "lack of software systems that are capable to enhance nominal mass spectral data files, that are flexible, extensible and that offer an easy to use graphical user interface." The group concluded that "OpenChrom will be hopefully extended by contributing developers, scientists and companies in the future." As of 2016, development on OpenChrom does indeed continue, with a 1.1.0 preview release available released in February.
 This 2012 paper, published in Journal of Pathology Informatics, provides the perspective of Yale University School of Medicine's Pathology Informatics Unit in regards to their experience with custom software development. Despite perceptions concerning the pitfalls of laboratories developing their own software, Sinard et al. conclude that "[m]any of the risks associated with custom development can be mitigated by a well-structured development process, use of open-source tools, and embracing an agile development philosophy."
This 2012 paper, published in Journal of Pathology Informatics, provides the perspective of Yale University School of Medicine's Pathology Informatics Unit in regards to their experience with custom software development. Despite perceptions concerning the pitfalls of laboratories developing their own software, Sinard et al. conclude that "[m]any of the risks associated with custom development can be mitigated by a well-structured development process, use of open-source tools, and embracing an agile development philosophy."
 Published in 2012, this paper on the NG6 next generation sequencing (NGS) platform describes the software's development process at GenoToul, a French shared life sciences research facility. Mariette et al. found that NGS software Galaxy was promising but not sufficient as it only "aims at simplifying data processing for researchers," whereas the group needed a LIMS that was also good at "gathering specialized pipelines and website" data. NG6 was developed to meet that need.
Published in 2012, this paper on the NG6 next generation sequencing (NGS) platform describes the software's development process at GenoToul, a French shared life sciences research facility. Mariette et al. found that NGS software Galaxy was promising but not sufficient as it only "aims at simplifying data processing for researchers," whereas the group needed a LIMS that was also good at "gathering specialized pipelines and website" data. NG6 was developed to meet that need.
 Hernández-de-Diego et al. from the Centro de Investigación Príncipe Felipe and the Karolinska Institute needed a softwar that " [i]n contrast to other solutions that put the focus on management of thousands of samples for core sequencing facilities ... [could handle] the annotation of experiments designed and run at individual research laboratories." Additionally, genomics solutions such as BASE had annotation limitations in regards to microarray experiment data. The group decided to develop their own system called the STATegra experiment management system (EMS), which they concluded "provides an integrated system for annotation of complex high-throughput omics experiments at functional genomics research laboratories."
Hernández-de-Diego et al. from the Centro de Investigación Príncipe Felipe and the Karolinska Institute needed a softwar that " [i]n contrast to other solutions that put the focus on management of thousands of samples for core sequencing facilities ... [could handle] the annotation of experiments designed and run at individual research laboratories." Additionally, genomics solutions such as BASE had annotation limitations in regards to microarray experiment data. The group decided to develop their own system called the STATegra experiment management system (EMS), which they concluded "provides an integrated system for annotation of complex high-throughput omics experiments at functional genomics research laboratories."
 In this 2012 journal article from ZooKeys, Blagoderov and his colleagues from the Natural History Museum in London describe the process they went through to improve the mass-digitization of their numerous collections. Noting problems with cost, "as well as the inherent fragmentation in collection based biodiversity informatics," the researchers created a "wall-to-wall" process using the SatScan tray scanning system. The group suggested that such a system could, when implemented well, "open up collections to the world, facilitating their use, and help create a global collection index that can be used to set priorities for further digitization."
In this 2012 journal article from ZooKeys, Blagoderov and his colleagues from the Natural History Museum in London describe the process they went through to improve the mass-digitization of their numerous collections. Noting problems with cost, "as well as the inherent fragmentation in collection based biodiversity informatics," the researchers created a "wall-to-wall" process using the SatScan tray scanning system. The group suggested that such a system could, when implemented well, "open up collections to the world, facilitating their use, and help create a global collection index that can be used to set priorities for further digitization."
 Reilly et al. at the University of North Carolina needed a specific solution for their genomics research, a solution that would "address the organizational challenges of computation-intensive biomedical research within a decentralized academic institution." After reviewing numerous options, the team decided to create their own system, MaPSeq, an application built on service-oriented architecture. In this paper summing up their development and implementation process, the group concludes that while the software is useful for massively parallel sequencing, "the general architecture and approach can be adapted for other complex or computationally-intense workflows."
Reilly et al. at the University of North Carolina needed a specific solution for their genomics research, a solution that would "address the organizational challenges of computation-intensive biomedical research within a decentralized academic institution." After reviewing numerous options, the team decided to create their own system, MaPSeq, an application built on service-oriented architecture. In this paper summing up their development and implementation process, the group concludes that while the software is useful for massively parallel sequencing, "the general architecture and approach can be adapted for other complex or computationally-intense workflows."
 In this brief article published in Frontiers in Environmental Science in 2014, environmental researcher Alexander Kokhanovsky takes a look at the state of environmental informatics past and present while also speculating about the future challenges. He ends by emphasizing the need for improved "estimation of aerosol load using space-borne instrumentation" to improve environmental research, including improved algorithms and visualization tools.
In this brief article published in Frontiers in Environmental Science in 2014, environmental researcher Alexander Kokhanovsky takes a look at the state of environmental informatics past and present while also speculating about the future challenges. He ends by emphasizing the need for improved "estimation of aerosol load using space-borne instrumentation" to improve environmental research, including improved algorithms and visualization tools.
 van Gaans et al. of the University of South Australia and James Cook University needed a system that provides data management functionality for the specific needs of a public health research organization, including data sharing and reuse. They needed a system that would "ensure that: data that are unmanaged be managed, data that are disconnected be connected, data that are invisible be findable, [and] data that are single use be reusable, within a structured collection." In this journal article, the group documents the development of their answer: the Public Health Data Management System (PHRDMS).
van Gaans et al. of the University of South Australia and James Cook University needed a system that provides data management functionality for the specific needs of a public health research organization, including data sharing and reuse. They needed a system that would "ensure that: data that are unmanaged be managed, data that are disconnected be connected, data that are invisible be findable, [and] data that are single use be reusable, within a structured collection." In this journal article, the group documents the development of their answer: the Public Health Data Management System (PHRDMS).
 Nanomedicine researchers at the University of Southern Denmark required a "dedicated LIMS for the management of large vector construct and cell line libraries." With no other open-source options available, the team developed their own, OpenLabFramework. Documenting their process, List et al. conclude "OLF can be deployed using different database management systems either locally, to a server, or to the cloud," and "[t]he incorporation of modern technologies, such as mobile devices and printing of barcode labels may increase productivity even further."
Nanomedicine researchers at the University of Southern Denmark required a "dedicated LIMS for the management of large vector construct and cell line libraries." With no other open-source options available, the team developed their own, OpenLabFramework. Documenting their process, List et al. conclude "OLF can be deployed using different database management systems either locally, to a server, or to the cloud," and "[t]he incorporation of modern technologies, such as mobile devices and printing of barcode labels may increase productivity even further."
 In this research paper, Nguyen et al. of Monash University in Australia "describes the design, implementation and operation of a multi-modality research imaging data management system that manages imaging data obtained from biomedical imaging scanners" at their facility. Faced with limitations of existing image management software and frameworks, the group custom built a system "based on DaRIS and XNAT has been designed and implemented to enable researchers to acquire, manage and analyse large, longitudinal biomedical imaging datasets."
In this research paper, Nguyen et al. of Monash University in Australia "describes the design, implementation and operation of a multi-modality research imaging data management system that manages imaging data obtained from biomedical imaging scanners" at their facility. Faced with limitations of existing image management software and frameworks, the group custom built a system "based on DaRIS and XNAT has been designed and implemented to enable researchers to acquire, manage and analyse large, longitudinal biomedical imaging datasets."
 Stahl et al. and Universités de Montpellier needed a system that could "streamline [biological] data storage and annotation collaboratively." Not finding a system to their liking, the group developed a Joomla!-based LIMS called Djeen and published their research on the development process in 2013. The group concludes: "Djeen allows managing project associated with heterogeneous data types while enforcing annotation integrity and minimum information. Projects are managed within a hierarchy and user permissions are finely-grained for each project, user and group."
Stahl et al. and Universités de Montpellier needed a system that could "streamline [biological] data storage and annotation collaboratively." Not finding a system to their liking, the group developed a Joomla!-based LIMS called Djeen and published their research on the development process in 2013. The group concludes: "Djeen allows managing project associated with heterogeneous data types while enforcing annotation integrity and minimum information. Projects are managed within a hierarchy and user permissions are finely-grained for each project, user and group."
 In 2010, O'Connor et al. published a paper on their experience developing and implementing a cloud-based query engine that can support thousands of genome datasets. Still actively developed as of 2015, the SeqWare Query Engine "can load and query variants (SNVs, indels, translocations, etc) with a rich level of annotations including coverage and functional consequences." The research team concluded in their paper that the software was then "capable of supporting large scale genome sequencing research projects involving hundreds to thousands of genomes as well as future large scale clinical deployments utilizing advanced sequencer technology that will soon involve tens to hundreds of thousands of genomes."
In 2010, O'Connor et al. published a paper on their experience developing and implementing a cloud-based query engine that can support thousands of genome datasets. Still actively developed as of 2015, the SeqWare Query Engine "can load and query variants (SNVs, indels, translocations, etc) with a rich level of annotations including coverage and functional consequences." The research team concluded in their paper that the software was then "capable of supporting large scale genome sequencing research projects involving hundreds to thousands of genomes as well as future large scale clinical deployments utilizing advanced sequencer technology that will soon involve tens to hundreds of thousands of genomes."
 In 2015, Singh et al. published their notes and reflections on developing SaDA, designed "for storing, retrieving and analyzing data originated from microorganism monitoring experiments." The group developed the software after discovering a lack of free, open-source software for microarray data management and analysis. The group concluded that "the platform has the potential to become an appropriate tool for a wide range of users focused not only in water based environmental research but also in other studies aimed at exploring and analyzing complex ecological habitats."
In 2015, Singh et al. published their notes and reflections on developing SaDA, designed "for storing, retrieving and analyzing data originated from microorganism monitoring experiments." The group developed the software after discovering a lack of free, open-source software for microarray data management and analysis. The group concluded that "the platform has the potential to become an appropriate tool for a wide range of users focused not only in water based environmental research but also in other studies aimed at exploring and analyzing complex ecological habitats."
 Seeking to overcome some of the challenges of massively parallel DNA sequencing, Reid et al. developed the Mercury analysis pipeline and deployed it on Amazon Web Services. Publishing their results in 2014 in BMC Bioinformatics, the group demonstrated the success of "a powerful combination of a robust and fully validated software pipeline and a scalable computational resource that, to date, we have applied to more than 10,000 whole genome and whole exome samples."
Seeking to overcome some of the challenges of massively parallel DNA sequencing, Reid et al. developed the Mercury analysis pipeline and deployed it on Amazon Web Services. Publishing their results in 2014 in BMC Bioinformatics, the group demonstrated the success of "a powerful combination of a robust and fully validated software pipeline and a scalable computational resource that, to date, we have applied to more than 10,000 whole genome and whole exome samples."
 Appearing in the magazine Open Source Business Resource (today called Technology Innovation Management Review) in 2011, this non-journal article by Sandro Groganz describes how "[o]pen source vendors can benefit from business ecosystems that form around their products," using open-source OXID eShop and its vendor OXID eSales as a representative example. Groganz concludes that "an open source community is not a state but a process," one that "creates a foundation for long-term growth and sustainability" when appropriately embraced.
Appearing in the magazine Open Source Business Resource (today called Technology Innovation Management Review) in 2011, this non-journal article by Sandro Groganz describes how "[o]pen source vendors can benefit from business ecosystems that form around their products," using open-source OXID eShop and its vendor OXID eSales as a representative example. Groganz concludes that "an open source community is not a state but a process," one that "creates a foundation for long-term growth and sustainability" when appropriately embraced.
 The San Raffaele Telethon Institute for Gene Therapy wanted a new LIMS to manage the data coming from their PCR techniques and next generation sequencing (NGS) methods. Not finding something suitable, the group developed its own LIMS, adLIMS. This 2015 research paper by Calabria et al., published in BMC Bioinformatics, covers the academic aspects of the information collection and development process.
The San Raffaele Telethon Institute for Gene Therapy wanted a new LIMS to manage the data coming from their PCR techniques and next generation sequencing (NGS) methods. Not finding something suitable, the group developed its own LIMS, adLIMS. This 2015 research paper by Calabria et al., published in BMC Bioinformatics, covers the academic aspects of the information collection and development process.
 This 2014 research published in BMC Bioinformatics sees Grimes and Ji presenting their "highly configurable and extensible" web-based laboratory information management system (LIMS) for next generation DNA sequencing, MendeLIMS. The group concludes that the software is "an invaluable tool for the management of our clinical sequencing studies," primarily due to its ability to reduce sample tracking errors and give "a comprehensive view of data being sequenced."
This 2014 research published in BMC Bioinformatics sees Grimes and Ji presenting their "highly configurable and extensible" web-based laboratory information management system (LIMS) for next generation DNA sequencing, MendeLIMS. The group concludes that the software is "an invaluable tool for the management of our clinical sequencing studies," primarily due to its ability to reduce sample tracking errors and give "a comprehensive view of data being sequenced."
 In this 2014 article published in BMC Bioinformatics, Dander et al., not content with the disparity among existing cancer treatment and bioinformatics platforms, discussed the results of creating Personalized Oncology Suite (POS). The web-based, scalable software system "integrates clinical data, NGS data and whole-slide bioimages from tissue sections" and, as the team concludes, "can be used not only in the context of cancer immunology but also in other studies in which NGS data and images of tissue sections are generated."
In this 2014 article published in BMC Bioinformatics, Dander et al., not content with the disparity among existing cancer treatment and bioinformatics platforms, discussed the results of creating Personalized Oncology Suite (POS). The web-based, scalable software system "integrates clinical data, NGS data and whole-slide bioimages from tissue sections" and, as the team concludes, "can be used not only in the context of cancer immunology but also in other studies in which NGS data and images of tissue sections are generated."















