Posted on August 1, 2016
By John Jones
Journal articles
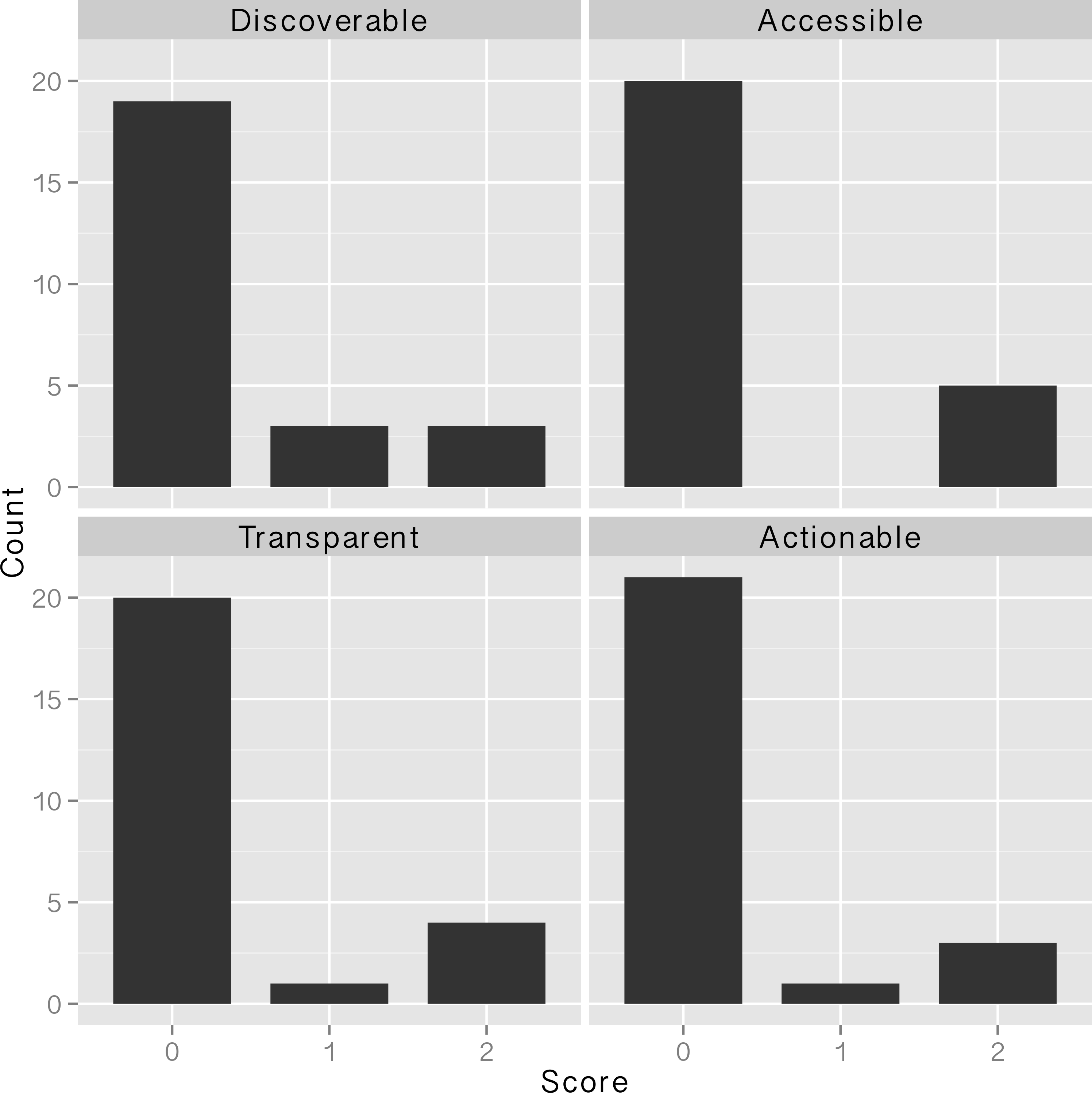
In this 2016 article published in
PLOS ONE, Van Tuyl and Whitmire take a close look at what "data sharing" means and what data sharing practices researchers have been using since the National Science Foundation's data management plan (DMP) requirements went into effect in 2011. Making federally-funded research "data functional for reuse, validation, meta-analysis, and replication of research" should be priority, they argue; however, they conclude not enough is being done in general. The researchers close by making "simple recommendations to data producers, publishers, repositories, and funding agencies that [they] believe will support more effective data sharing."
Posted on July 25, 2016
By John Jones
Journal articles
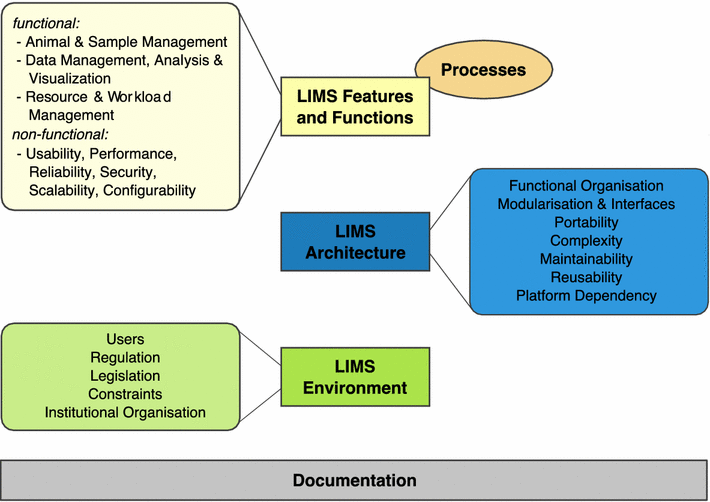
Clinical researchers conducting mouse studies at seven different facilities around the world shared their experiences using a laboratory information management system (LIMS) in order "to facilitate or even enable mouse and data management" better in their facilities. This 2015 paper by Maier
et al. examines those discussions and final findings in a "review" format, concluding "the unique LIMS environment in a particular facility strongly influences strategic LIMS decisions and LIMS development" though "there is no universal LIMS for the mouse research domain that fits all requirements."
Posted on July 19, 2016
By John Jones
Journal articles
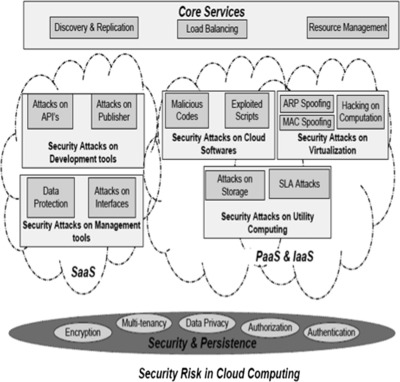
In this 2016 article in
Applied Computing and Informatics, Hussain
et al. take a closer look at the types of security attacks specific to cloud-based offerings and proposes a new multi-level classification model to clarify them, with an end goal "to determine the risk level and type of security required for each service at different cloud layers for a cloud consumer and cloud provider."
Posted on July 12, 2016
By John Jones
Journal articles

Published in the
Journal of eScience Librarianship, this 2016 article by Norton
et al. looks at the topic of "big data" management in clinical and translational research from the university and library standpoint. As academic libraries are a major component of such research, Norton
et al. reached out to the various medical colleges at the University of Florida and sought to clarify researcher needs. The group concludes that its research has led to "addressing common campus-wide concerns through data management training, collaboration with campus IT infrastructure and research units, and creating a Data Management Librarian position" to improve the library system's role with data management for clinical researchers.
Posted on July 6, 2016
By John Jones
Journal articles
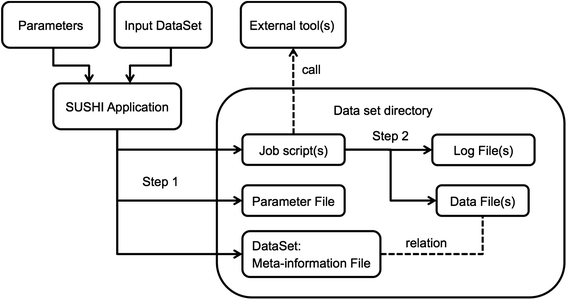
Many next-generation sequencing (NGS) data analysis frameworks exist, from Galaxy to bpipe. However, Hatakeyama
et al. at the University of Zürich noted a distinct lack of a framework that 1. offers both web-based and scripting options and 2. "puts an emphasis on having a human-readable and portable file-based representation of the meta-information and associated data." In response, the researchers created SUSHI (Support Users for SHell-script Integration). They conclude that "[i]n one solution, SUSHI provides at the same time fully documented, high level NGS analysis tools to biologists and an easy to administer, reproducible approach for large and complicated NGS data to bioinformaticians."
Posted on June 30, 2016
By John Jones
Journal articles
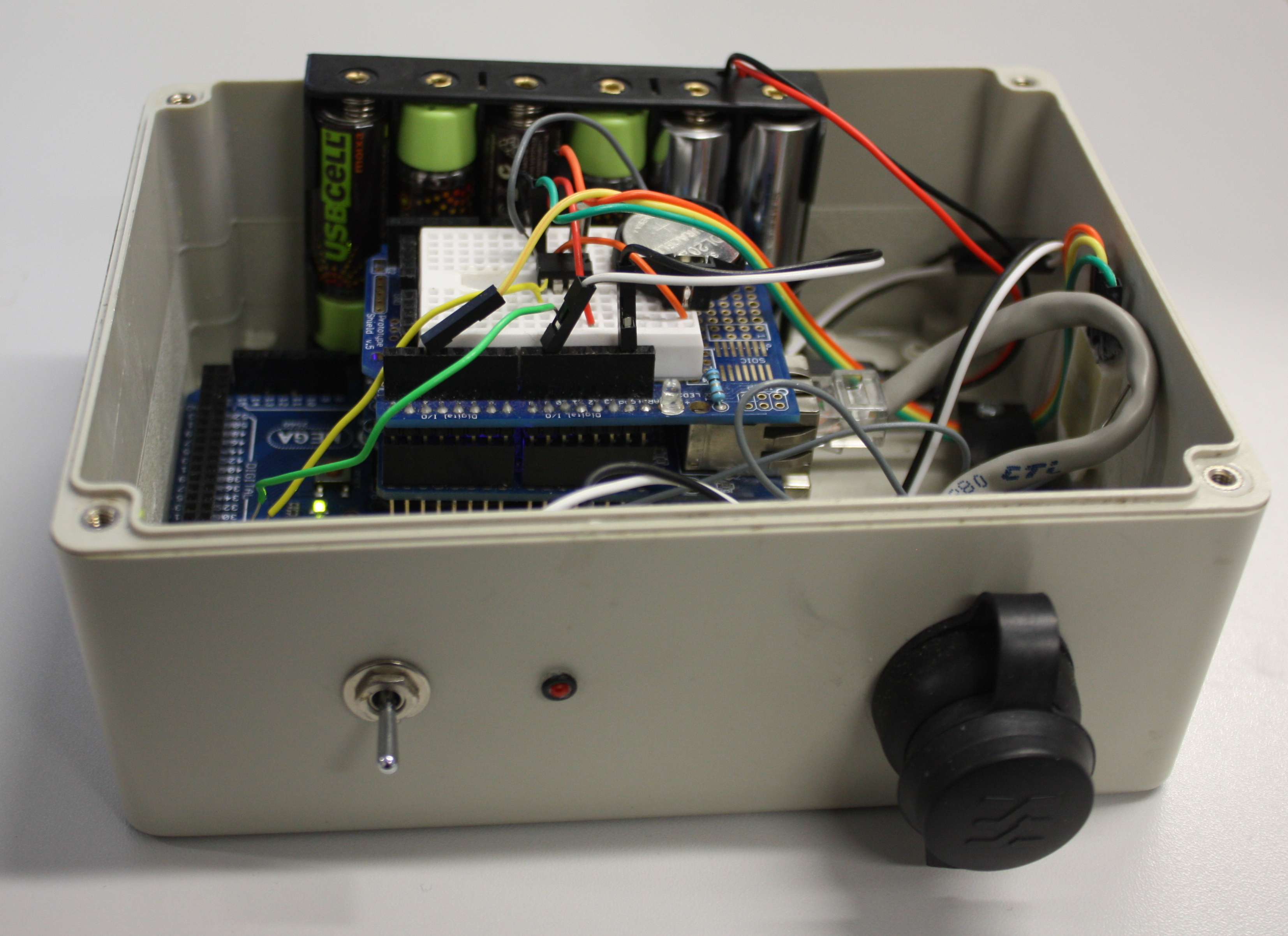
This 2014 paper by Ed Baker of London's Natural History Museum outlines a methodology for combining open-source software such as Drupal with open hardware like Arduino to create a real-time environmental monitoring station that is low-power and low-cost. Baker outlines step by step his approach (he calls it a "how to guide") to creating an open-source environmental data logger that incorporates a digital temperature and humidity sensor. Though he offers no formal conclusions, Baker states: "It is hoped that the publication of this device will encourage biodiversity scientists to collaborate outside of their discipline, whether it be with citizen engineers or professional academics."
Posted on June 21, 2016
By John Jones
Journal articles
Evaluating health information systems/technology is no easy task. Eivazzadeh
et al. recognize that, as well as the fact that developing evaluation frameworks presents its own set of challenges. Having looked at several different models, the researchers wished to develop their own evaluation method, one that taps into "evaluation aspects for a set of one or more health information systems — whether similar or heterogeneous — by organizing, unifying, and aggregating the quality attributes extracted from those systems and from an external evaluation framework." As such, the group developed the UVON method, which they conclude can be used " to create ontologies for evaluation" of health information systems as well as "to mix them with elements from other evaluation frameworks."
Posted on June 14, 2016
By John Jones
Journal articles
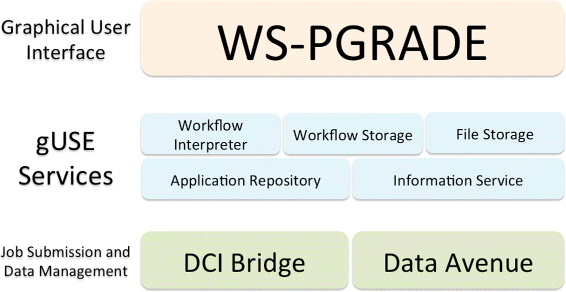
This featured article from the journal
BMC Bioinformatics falls on the heels of several years of discussion on the topic of reproducibility of a scientific experiment's end results. De la Garza
et al. point to workflows and their repeatability as vital cogs in such efforts. "Breaking down the complexity of such experiments into the joint collaboration of small, repeatable, well defined tasks, each with well defined inputs, parameters, and outputs, offers the immediate benefit of identifying bottlenecks, pinpoint sections which could benefit from parallelization," they state. The researchers developed their own set of free platform-independent tools for designing, executing, and sharing workflows. They conclude: "We are confident that our work presented in this document ... not only provides scientists a way to design and test workflows on their desktop computers, but also enables them to use powerful resources to execute their workflows, thus producing scientific results in a timely manner."
Posted on June 7, 2016
By John Jones
Journal articles
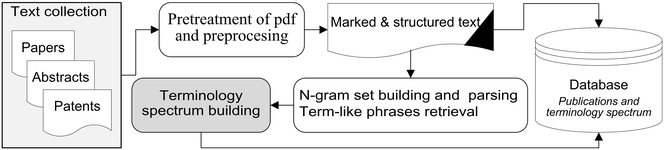
In this 2016 journal article published in
Journal of Cheminformatics, Alperin
et al. present the fruits of their labor in an attempt to " to develop, test and assess a methodology" for both extracting and categorizing words and terminology from chemistry-related PDFs, with the goal of being able to apply "textual analysis across document collections." They conclude that "[t]erminology spectrum retrieval may be used to perform various types of text analysis across document collections" as well as "to find out research trends and new concepts in the subject field by registering changes in terminology usage in the most rapidly developing areas of research."
Posted on June 3, 2016
By John Jones
Journal articles

Digital health services is an expanding force, empowering people to track and manage their health. However, it comes with cost and legal concerns, requiring a legal framework for the development and assessment of those services. In this 2016 paper appearing in
JMIR Medical Informatics, Garrell
et al. lay out such a framework based around Swedish law, though leaving room for the framework to be adapted to other regions of the world. They conclude that their framework "can be used in prospective evaluation of the relationship of a potential health-promoting digital service with the existing laws and regulations" of a particular region.
Posted on May 25, 2016
By John Jones
Journal articles
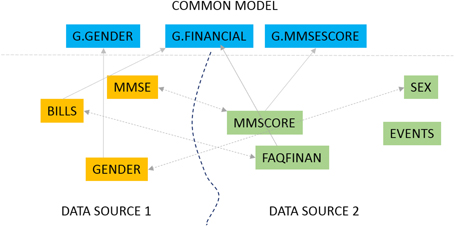
In this 2016 article appearing in
Frontiers in Neuroinformatics, Ashish
et al. present GEM, "an intelligent software assistant for automated data mapping across different datasets or from a dataset to a common data model." Used for Alzheimer research though applicable to many other fields, the group concludes "[o]ur experimental evaluations demonstrate significant mapping accuracy improvements obtained with our approach, particularly by leveraging the detailed information synthesized for data dictionaries."
Posted on May 16, 2016
By John Jones
Journal articles
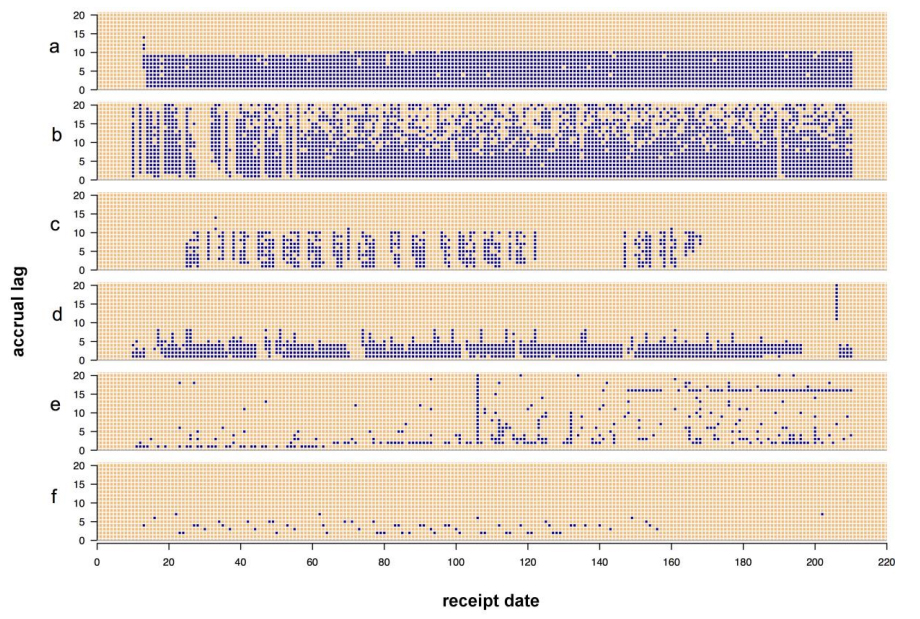
The state of data management across the sciences is getting increasingly complex as data stores build up, and the world of public health is no less affected. Making sense of data is one portion of management, but quality analysis is also an important but slightly understated aspect as well. This 2015 paper by Eaton
et al. explains a series of "data quality tools developed to gain insight into the data quality problems associated with these data." The group concludes "our key insight was the need to assess temporal patterns in the data in terms of accrual lag."
Posted on May 9, 2016
By John Jones
Journal articles
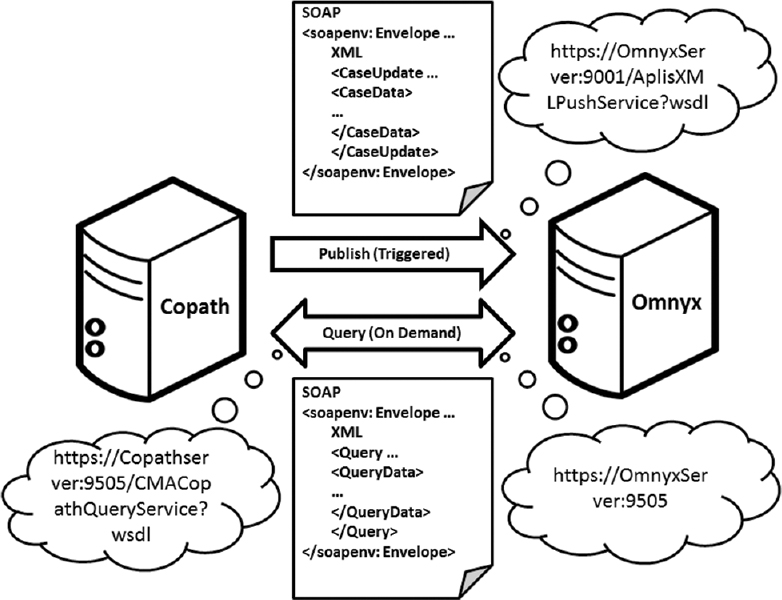
What happens when you integrate a digital pathology system (DPS) with an anatomical pathology laboratory information system (APLIS)? In the case of Guo et al. and the University of Pittsburgh Medical Center, "[t]he integration streamlined our digital sign-out workflow, diminished the potential for human error related to matching slides, and improved the sign-out experience for pathologists." This paper, published in
Journal of Pathology Informatics in 2016, describes their line of thinking, integration plans, and final results.
Posted on May 5, 2016
By John Jones
Journal articles
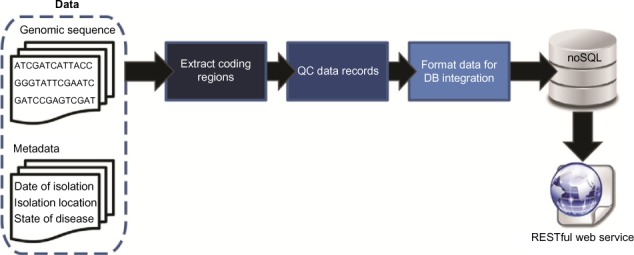
In this 2016 paper published in
Evolutionary Bioinformatics, Reisman
et al. discuss a polyglot approach "involving multiple languages, libraries, and persistence mechanisms" towards managing genomic sequence data. Using a NoSQL and RESTful web service approach, the team tested their developed pipeline on an evolutionary study of HIV-1. They conclude that " the case study highlights the abilities of the tool," and "although utilized for the investigation of a virus here, the approach can be applied to any species of interest."
Posted on April 25, 2016
By John Jones
Journal articles
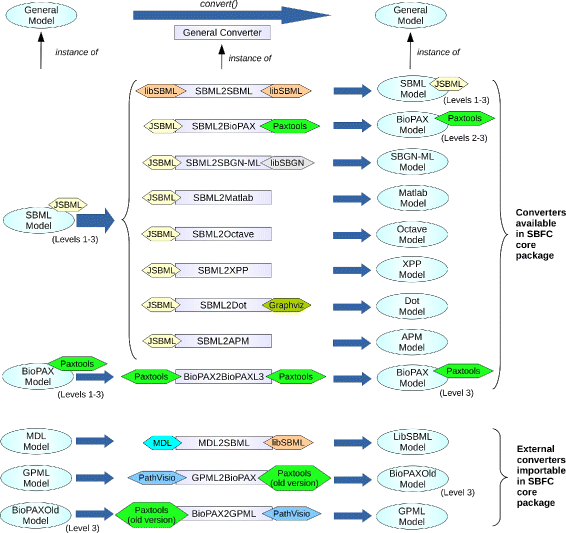
Rodriguez
et al. found that when converting computational models from one format to another, while many tools exist, they tend not to be very interoperable and can often be redundant. Additionally, they can be unmaintained or left abandoned. The researchers saw a need for a modular, open-source software system "to support rapid implementation and integration of new converters" in a more collaboratory way. They developed the System Biology Format Converter (SBFC), a Java-based tool that, per their conclusion, "helps computational biologists to process or visualise their models using different software tools, and software developers to implement format conversion."
Posted on April 19, 2016
By John Jones
Journal articles
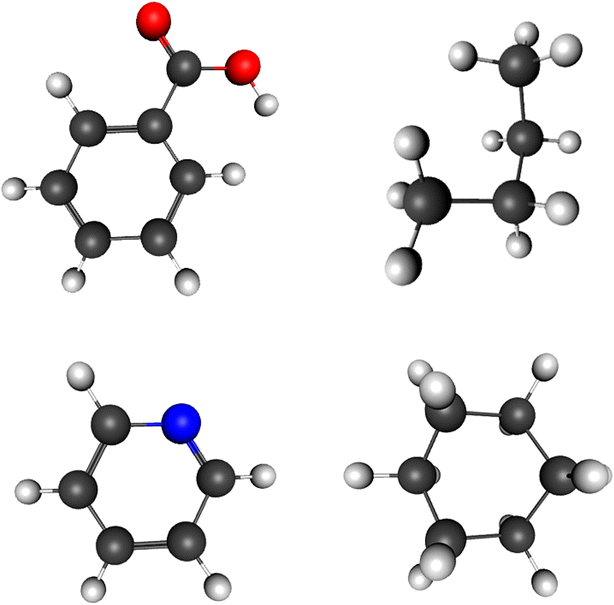
In this 2015 journal article published in the open-access journal
Journal of Cheminformatics, Mohebifar and Sajadi describe their web-based HTML5/CSS3 3D molecule editor and visualizer Chemozart. Able to be run from the public web source or your own personal instance, Chemozart is both useful for educational and research purposes. The authors tout "that there’s no need to install anything and it can be accessed easily via a URL."
Posted on April 13, 2016
By John Jones
Journal articles
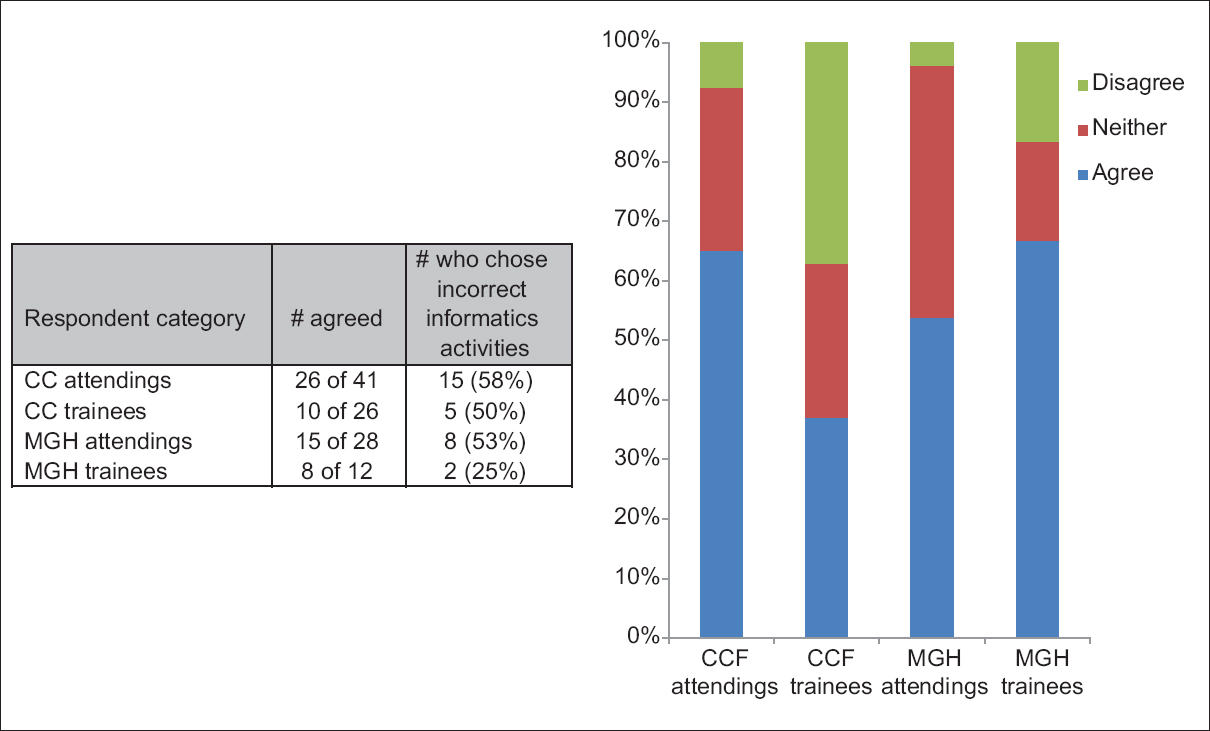
Perhaps frustrated with the state of education and misperceptions in regards to pathology informatics (PI), Walker
et al. set out to conduct a survey of noninformatics-oriented pathologists and trainees at the Cleveland Clinic and Massachusetts General Hospital to better grasp views of the professional field. In this paper published in
Journal of Pathology Informatics in April 2016, the researchers present their findings and opine about the state of pathology informatics education. They conclude: "Improved understanding and acceptance of PI throughout the pathology community could facilitate the communication and cooperation necessary to realize the type of informatics initiatives capable of advancing the importance of pathologists in the changing healthcare environment."
Posted on April 6, 2016
By John Jones
Journal articles

In this "opinion article" published in the open-access journal
F1000Research, Dirnagl and Przesdzing argue for the benefits of — and recognize the occasional problems with — electronic laboratory notebooks (ELNs). The duo attempts to answer the questions "What does it afford you, what does it require, and how should you go about implementing it?" They conclude that ELNs improve workflow and collaboration, allow for greater data integration, reduce transcription errors, improve data quality, and promote compliance with a variety of practices and regulations. "We have no doubt that ELNs will become standard in most life science laboratories in the near future," they state at the end.
Posted on April 2, 2016
By John Jones
Journal articles
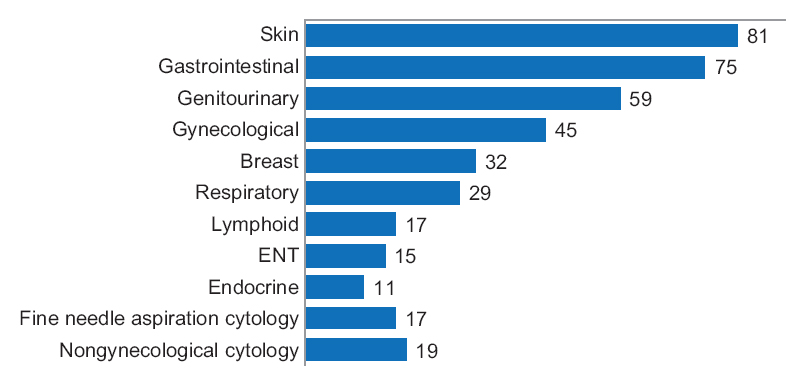
Is digital diagnostic determination of pathology images quicker than microscopic? It depends on who you ask. Vodovnik published results of his comparative study in early 2016, comparing digital and microscopic diagnostic times using tools such as digital pathology workstations and laboratory information management systems. His conclusion: "A shorter diagnostic time in digital pathology comparing with traditional microscopy can be achieved in the routine diagnostic setting with adequate and stable network speeds, fully integrated LIMS and double displays as default parameters, in addition to better ergonomics, larger viewing field, and absence of physical slide handling, with effects on the both diagnostic and nondiagnostic time."
Posted on March 26, 2016
By John Jones
Journal articles
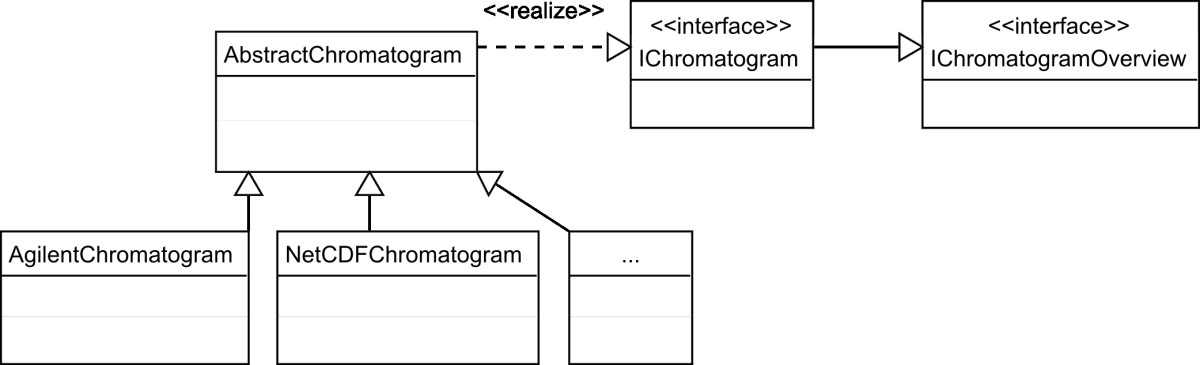
In 2010, Philip Wenig and Juergen Odermatt published their experiences on developing OpenChrom, an open-source chromatography data management system (CDMS) for analyzing mass spectrometric chromatographic data. The team built the software due to a perceived "lack of software systems that are capable to enhance nominal mass spectral data files, that are flexible, extensible and that offer an easy to use graphical user interface." The group concluded that "OpenChrom will be hopefully extended by contributing developers, scientists and companies in the future." As of 2016, development on OpenChrom does indeed continue, with a 1.1.0 preview release available released in February.
 In this 2016 article published in PLOS ONE, Van Tuyl and Whitmire take a close look at what "data sharing" means and what data sharing practices researchers have been using since the National Science Foundation's data management plan (DMP) requirements went into effect in 2011. Making federally-funded research "data functional for reuse, validation, meta-analysis, and replication of research" should be priority, they argue; however, they conclude not enough is being done in general. The researchers close by making "simple recommendations to data producers, publishers, repositories, and funding agencies that [they] believe will support more effective data sharing."
In this 2016 article published in PLOS ONE, Van Tuyl and Whitmire take a close look at what "data sharing" means and what data sharing practices researchers have been using since the National Science Foundation's data management plan (DMP) requirements went into effect in 2011. Making federally-funded research "data functional for reuse, validation, meta-analysis, and replication of research" should be priority, they argue; however, they conclude not enough is being done in general. The researchers close by making "simple recommendations to data producers, publishers, repositories, and funding agencies that [they] believe will support more effective data sharing."
 Clinical researchers conducting mouse studies at seven different facilities around the world shared their experiences using a laboratory information management system (LIMS) in order "to facilitate or even enable mouse and data management" better in their facilities. This 2015 paper by Maier et al. examines those discussions and final findings in a "review" format, concluding "the unique LIMS environment in a particular facility strongly influences strategic LIMS decisions and LIMS development" though "there is no universal LIMS for the mouse research domain that fits all requirements."
Clinical researchers conducting mouse studies at seven different facilities around the world shared their experiences using a laboratory information management system (LIMS) in order "to facilitate or even enable mouse and data management" better in their facilities. This 2015 paper by Maier et al. examines those discussions and final findings in a "review" format, concluding "the unique LIMS environment in a particular facility strongly influences strategic LIMS decisions and LIMS development" though "there is no universal LIMS for the mouse research domain that fits all requirements."
 In this 2016 article in Applied Computing and Informatics, Hussain et al. take a closer look at the types of security attacks specific to cloud-based offerings and proposes a new multi-level classification model to clarify them, with an end goal "to determine the risk level and type of security required for each service at different cloud layers for a cloud consumer and cloud provider."
In this 2016 article in Applied Computing and Informatics, Hussain et al. take a closer look at the types of security attacks specific to cloud-based offerings and proposes a new multi-level classification model to clarify them, with an end goal "to determine the risk level and type of security required for each service at different cloud layers for a cloud consumer and cloud provider."
 Published in the Journal of eScience Librarianship, this 2016 article by Norton et al. looks at the topic of "big data" management in clinical and translational research from the university and library standpoint. As academic libraries are a major component of such research, Norton et al. reached out to the various medical colleges at the University of Florida and sought to clarify researcher needs. The group concludes that its research has led to "addressing common campus-wide concerns through data management training, collaboration with campus IT infrastructure and research units, and creating a Data Management Librarian position" to improve the library system's role with data management for clinical researchers.
Published in the Journal of eScience Librarianship, this 2016 article by Norton et al. looks at the topic of "big data" management in clinical and translational research from the university and library standpoint. As academic libraries are a major component of such research, Norton et al. reached out to the various medical colleges at the University of Florida and sought to clarify researcher needs. The group concludes that its research has led to "addressing common campus-wide concerns through data management training, collaboration with campus IT infrastructure and research units, and creating a Data Management Librarian position" to improve the library system's role with data management for clinical researchers.
 Many next-generation sequencing (NGS) data analysis frameworks exist, from Galaxy to bpipe. However, Hatakeyama et al. at the University of Zürich noted a distinct lack of a framework that 1. offers both web-based and scripting options and 2. "puts an emphasis on having a human-readable and portable file-based representation of the meta-information and associated data." In response, the researchers created SUSHI (Support Users for SHell-script Integration). They conclude that "[i]n one solution, SUSHI provides at the same time fully documented, high level NGS analysis tools to biologists and an easy to administer, reproducible approach for large and complicated NGS data to bioinformaticians."
Many next-generation sequencing (NGS) data analysis frameworks exist, from Galaxy to bpipe. However, Hatakeyama et al. at the University of Zürich noted a distinct lack of a framework that 1. offers both web-based and scripting options and 2. "puts an emphasis on having a human-readable and portable file-based representation of the meta-information and associated data." In response, the researchers created SUSHI (Support Users for SHell-script Integration). They conclude that "[i]n one solution, SUSHI provides at the same time fully documented, high level NGS analysis tools to biologists and an easy to administer, reproducible approach for large and complicated NGS data to bioinformaticians."
 This 2014 paper by Ed Baker of London's Natural History Museum outlines a methodology for combining open-source software such as Drupal with open hardware like Arduino to create a real-time environmental monitoring station that is low-power and low-cost. Baker outlines step by step his approach (he calls it a "how to guide") to creating an open-source environmental data logger that incorporates a digital temperature and humidity sensor. Though he offers no formal conclusions, Baker states: "It is hoped that the publication of this device will encourage biodiversity scientists to collaborate outside of their discipline, whether it be with citizen engineers or professional academics."
This 2014 paper by Ed Baker of London's Natural History Museum outlines a methodology for combining open-source software such as Drupal with open hardware like Arduino to create a real-time environmental monitoring station that is low-power and low-cost. Baker outlines step by step his approach (he calls it a "how to guide") to creating an open-source environmental data logger that incorporates a digital temperature and humidity sensor. Though he offers no formal conclusions, Baker states: "It is hoped that the publication of this device will encourage biodiversity scientists to collaborate outside of their discipline, whether it be with citizen engineers or professional academics."
 Evaluating health information systems/technology is no easy task. Eivazzadeh et al. recognize that, as well as the fact that developing evaluation frameworks presents its own set of challenges. Having looked at several different models, the researchers wished to develop their own evaluation method, one that taps into "evaluation aspects for a set of one or more health information systems — whether similar or heterogeneous — by organizing, unifying, and aggregating the quality attributes extracted from those systems and from an external evaluation framework." As such, the group developed the UVON method, which they conclude can be used " to create ontologies for evaluation" of health information systems as well as "to mix them with elements from other evaluation frameworks."
Evaluating health information systems/technology is no easy task. Eivazzadeh et al. recognize that, as well as the fact that developing evaluation frameworks presents its own set of challenges. Having looked at several different models, the researchers wished to develop their own evaluation method, one that taps into "evaluation aspects for a set of one or more health information systems — whether similar or heterogeneous — by organizing, unifying, and aggregating the quality attributes extracted from those systems and from an external evaluation framework." As such, the group developed the UVON method, which they conclude can be used " to create ontologies for evaluation" of health information systems as well as "to mix them with elements from other evaluation frameworks."
 This featured article from the journal BMC Bioinformatics falls on the heels of several years of discussion on the topic of reproducibility of a scientific experiment's end results. De la Garza et al. point to workflows and their repeatability as vital cogs in such efforts. "Breaking down the complexity of such experiments into the joint collaboration of small, repeatable, well defined tasks, each with well defined inputs, parameters, and outputs, offers the immediate benefit of identifying bottlenecks, pinpoint sections which could benefit from parallelization," they state. The researchers developed their own set of free platform-independent tools for designing, executing, and sharing workflows. They conclude: "We are confident that our work presented in this document ... not only provides scientists a way to design and test workflows on their desktop computers, but also enables them to use powerful resources to execute their workflows, thus producing scientific results in a timely manner."
This featured article from the journal BMC Bioinformatics falls on the heels of several years of discussion on the topic of reproducibility of a scientific experiment's end results. De la Garza et al. point to workflows and their repeatability as vital cogs in such efforts. "Breaking down the complexity of such experiments into the joint collaboration of small, repeatable, well defined tasks, each with well defined inputs, parameters, and outputs, offers the immediate benefit of identifying bottlenecks, pinpoint sections which could benefit from parallelization," they state. The researchers developed their own set of free platform-independent tools for designing, executing, and sharing workflows. They conclude: "We are confident that our work presented in this document ... not only provides scientists a way to design and test workflows on their desktop computers, but also enables them to use powerful resources to execute their workflows, thus producing scientific results in a timely manner."
 In this 2016 journal article published in Journal of Cheminformatics, Alperin et al. present the fruits of their labor in an attempt to " to develop, test and assess a methodology" for both extracting and categorizing words and terminology from chemistry-related PDFs, with the goal of being able to apply "textual analysis across document collections." They conclude that "[t]erminology spectrum retrieval may be used to perform various types of text analysis across document collections" as well as "to find out research trends and new concepts in the subject field by registering changes in terminology usage in the most rapidly developing areas of research."
In this 2016 journal article published in Journal of Cheminformatics, Alperin et al. present the fruits of their labor in an attempt to " to develop, test and assess a methodology" for both extracting and categorizing words and terminology from chemistry-related PDFs, with the goal of being able to apply "textual analysis across document collections." They conclude that "[t]erminology spectrum retrieval may be used to perform various types of text analysis across document collections" as well as "to find out research trends and new concepts in the subject field by registering changes in terminology usage in the most rapidly developing areas of research."
 Digital health services is an expanding force, empowering people to track and manage their health. However, it comes with cost and legal concerns, requiring a legal framework for the development and assessment of those services. In this 2016 paper appearing in JMIR Medical Informatics, Garrell et al. lay out such a framework based around Swedish law, though leaving room for the framework to be adapted to other regions of the world. They conclude that their framework "can be used in prospective evaluation of the relationship of a potential health-promoting digital service with the existing laws and regulations" of a particular region.
Digital health services is an expanding force, empowering people to track and manage their health. However, it comes with cost and legal concerns, requiring a legal framework for the development and assessment of those services. In this 2016 paper appearing in JMIR Medical Informatics, Garrell et al. lay out such a framework based around Swedish law, though leaving room for the framework to be adapted to other regions of the world. They conclude that their framework "can be used in prospective evaluation of the relationship of a potential health-promoting digital service with the existing laws and regulations" of a particular region.
 In this 2016 article appearing in Frontiers in Neuroinformatics, Ashish et al. present GEM, "an intelligent software assistant for automated data mapping across different datasets or from a dataset to a common data model." Used for Alzheimer research though applicable to many other fields, the group concludes "[o]ur experimental evaluations demonstrate significant mapping accuracy improvements obtained with our approach, particularly by leveraging the detailed information synthesized for data dictionaries."
In this 2016 article appearing in Frontiers in Neuroinformatics, Ashish et al. present GEM, "an intelligent software assistant for automated data mapping across different datasets or from a dataset to a common data model." Used for Alzheimer research though applicable to many other fields, the group concludes "[o]ur experimental evaluations demonstrate significant mapping accuracy improvements obtained with our approach, particularly by leveraging the detailed information synthesized for data dictionaries."
 The state of data management across the sciences is getting increasingly complex as data stores build up, and the world of public health is no less affected. Making sense of data is one portion of management, but quality analysis is also an important but slightly understated aspect as well. This 2015 paper by Eaton et al. explains a series of "data quality tools developed to gain insight into the data quality problems associated with these data." The group concludes "our key insight was the need to assess temporal patterns in the data in terms of accrual lag."
The state of data management across the sciences is getting increasingly complex as data stores build up, and the world of public health is no less affected. Making sense of data is one portion of management, but quality analysis is also an important but slightly understated aspect as well. This 2015 paper by Eaton et al. explains a series of "data quality tools developed to gain insight into the data quality problems associated with these data." The group concludes "our key insight was the need to assess temporal patterns in the data in terms of accrual lag."
 What happens when you integrate a digital pathology system (DPS) with an anatomical pathology laboratory information system (APLIS)? In the case of Guo et al. and the University of Pittsburgh Medical Center, "[t]he integration streamlined our digital sign-out workflow, diminished the potential for human error related to matching slides, and improved the sign-out experience for pathologists." This paper, published in Journal of Pathology Informatics in 2016, describes their line of thinking, integration plans, and final results.
What happens when you integrate a digital pathology system (DPS) with an anatomical pathology laboratory information system (APLIS)? In the case of Guo et al. and the University of Pittsburgh Medical Center, "[t]he integration streamlined our digital sign-out workflow, diminished the potential for human error related to matching slides, and improved the sign-out experience for pathologists." This paper, published in Journal of Pathology Informatics in 2016, describes their line of thinking, integration plans, and final results.
 In this 2016 paper published in Evolutionary Bioinformatics, Reisman et al. discuss a polyglot approach "involving multiple languages, libraries, and persistence mechanisms" towards managing genomic sequence data. Using a NoSQL and RESTful web service approach, the team tested their developed pipeline on an evolutionary study of HIV-1. They conclude that " the case study highlights the abilities of the tool," and "although utilized for the investigation of a virus here, the approach can be applied to any species of interest."
In this 2016 paper published in Evolutionary Bioinformatics, Reisman et al. discuss a polyglot approach "involving multiple languages, libraries, and persistence mechanisms" towards managing genomic sequence data. Using a NoSQL and RESTful web service approach, the team tested their developed pipeline on an evolutionary study of HIV-1. They conclude that " the case study highlights the abilities of the tool," and "although utilized for the investigation of a virus here, the approach can be applied to any species of interest."
 Rodriguez et al. found that when converting computational models from one format to another, while many tools exist, they tend not to be very interoperable and can often be redundant. Additionally, they can be unmaintained or left abandoned. The researchers saw a need for a modular, open-source software system "to support rapid implementation and integration of new converters" in a more collaboratory way. They developed the System Biology Format Converter (SBFC), a Java-based tool that, per their conclusion, "helps computational biologists to process or visualise their models using different software tools, and software developers to implement format conversion."
Rodriguez et al. found that when converting computational models from one format to another, while many tools exist, they tend not to be very interoperable and can often be redundant. Additionally, they can be unmaintained or left abandoned. The researchers saw a need for a modular, open-source software system "to support rapid implementation and integration of new converters" in a more collaboratory way. They developed the System Biology Format Converter (SBFC), a Java-based tool that, per their conclusion, "helps computational biologists to process or visualise their models using different software tools, and software developers to implement format conversion."
 In this 2015 journal article published in the open-access journal Journal of Cheminformatics, Mohebifar and Sajadi describe their web-based HTML5/CSS3 3D molecule editor and visualizer Chemozart. Able to be run from the public web source or your own personal instance, Chemozart is both useful for educational and research purposes. The authors tout "that there’s no need to install anything and it can be accessed easily via a URL."
In this 2015 journal article published in the open-access journal Journal of Cheminformatics, Mohebifar and Sajadi describe their web-based HTML5/CSS3 3D molecule editor and visualizer Chemozart. Able to be run from the public web source or your own personal instance, Chemozart is both useful for educational and research purposes. The authors tout "that there’s no need to install anything and it can be accessed easily via a URL."
 Perhaps frustrated with the state of education and misperceptions in regards to pathology informatics (PI), Walker et al. set out to conduct a survey of noninformatics-oriented pathologists and trainees at the Cleveland Clinic and Massachusetts General Hospital to better grasp views of the professional field. In this paper published in Journal of Pathology Informatics in April 2016, the researchers present their findings and opine about the state of pathology informatics education. They conclude: "Improved understanding and acceptance of PI throughout the pathology community could facilitate the communication and cooperation necessary to realize the type of informatics initiatives capable of advancing the importance of pathologists in the changing healthcare environment."
Perhaps frustrated with the state of education and misperceptions in regards to pathology informatics (PI), Walker et al. set out to conduct a survey of noninformatics-oriented pathologists and trainees at the Cleveland Clinic and Massachusetts General Hospital to better grasp views of the professional field. In this paper published in Journal of Pathology Informatics in April 2016, the researchers present their findings and opine about the state of pathology informatics education. They conclude: "Improved understanding and acceptance of PI throughout the pathology community could facilitate the communication and cooperation necessary to realize the type of informatics initiatives capable of advancing the importance of pathologists in the changing healthcare environment."
 In this "opinion article" published in the open-access journal F1000Research, Dirnagl and Przesdzing argue for the benefits of — and recognize the occasional problems with — electronic laboratory notebooks (ELNs). The duo attempts to answer the questions "What does it afford you, what does it require, and how should you go about implementing it?" They conclude that ELNs improve workflow and collaboration, allow for greater data integration, reduce transcription errors, improve data quality, and promote compliance with a variety of practices and regulations. "We have no doubt that ELNs will become standard in most life science laboratories in the near future," they state at the end.
In this "opinion article" published in the open-access journal F1000Research, Dirnagl and Przesdzing argue for the benefits of — and recognize the occasional problems with — electronic laboratory notebooks (ELNs). The duo attempts to answer the questions "What does it afford you, what does it require, and how should you go about implementing it?" They conclude that ELNs improve workflow and collaboration, allow for greater data integration, reduce transcription errors, improve data quality, and promote compliance with a variety of practices and regulations. "We have no doubt that ELNs will become standard in most life science laboratories in the near future," they state at the end.
 Is digital diagnostic determination of pathology images quicker than microscopic? It depends on who you ask. Vodovnik published results of his comparative study in early 2016, comparing digital and microscopic diagnostic times using tools such as digital pathology workstations and laboratory information management systems. His conclusion: "A shorter diagnostic time in digital pathology comparing with traditional microscopy can be achieved in the routine diagnostic setting with adequate and stable network speeds, fully integrated LIMS and double displays as default parameters, in addition to better ergonomics, larger viewing field, and absence of physical slide handling, with effects on the both diagnostic and nondiagnostic time."
Is digital diagnostic determination of pathology images quicker than microscopic? It depends on who you ask. Vodovnik published results of his comparative study in early 2016, comparing digital and microscopic diagnostic times using tools such as digital pathology workstations and laboratory information management systems. His conclusion: "A shorter diagnostic time in digital pathology comparing with traditional microscopy can be achieved in the routine diagnostic setting with adequate and stable network speeds, fully integrated LIMS and double displays as default parameters, in addition to better ergonomics, larger viewing field, and absence of physical slide handling, with effects on the both diagnostic and nondiagnostic time."
 In 2010, Philip Wenig and Juergen Odermatt published their experiences on developing OpenChrom, an open-source chromatography data management system (CDMS) for analyzing mass spectrometric chromatographic data. The team built the software due to a perceived "lack of software systems that are capable to enhance nominal mass spectral data files, that are flexible, extensible and that offer an easy to use graphical user interface." The group concluded that "OpenChrom will be hopefully extended by contributing developers, scientists and companies in the future." As of 2016, development on OpenChrom does indeed continue, with a 1.1.0 preview release available released in February.
In 2010, Philip Wenig and Juergen Odermatt published their experiences on developing OpenChrom, an open-source chromatography data management system (CDMS) for analyzing mass spectrometric chromatographic data. The team built the software due to a perceived "lack of software systems that are capable to enhance nominal mass spectral data files, that are flexible, extensible and that offer an easy to use graphical user interface." The group concluded that "OpenChrom will be hopefully extended by contributing developers, scientists and companies in the future." As of 2016, development on OpenChrom does indeed continue, with a 1.1.0 preview release available released in February.
















