Infrastructure tools to support an effective radiation oncology learning health system
Original file (1,367 × 630 pixels, file size: 153 KB, MIME type: image/jpeg)
Summary
| Description |
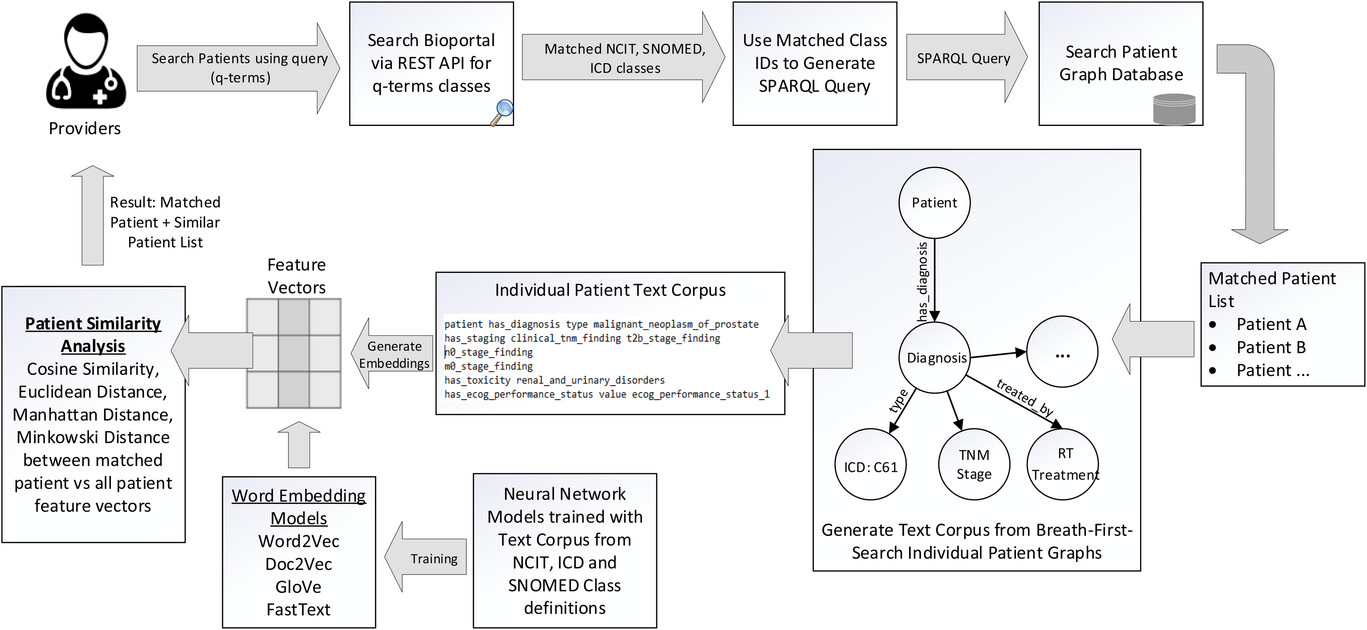
Figure 3. Design architecture for the ontology-based keyword search system. When the user wants to query the patient graph database to retrieve matching records, the only input necessary is the medical terms (q-terms) and an indication to include any synonym, parent, or children terminology classes in the search. The software queries the Bioportal API and retrieves all the matching NCIT, SNOMED, ICD-10 classes to the q-terms. A SPARQL query is generated and executed on the graph database SPARQL endpoint, and the results indicating the matching patient records and their corresponding data fields are displayed to the user. Our architecture includes the generation of text corpus from breadth-first search (BFS) of individual patient graphs and using word embedding models to generate feature vectors to identify similar patient cohorts. |
|---|---|
| Source |
Kapoor, Rishabh; Sleeman IV, William C.; Ghosh, Preetam; Palta, Jatinder (2023). "Infrastructure tools to support an effective radiation oncology learning health system". Journal of Applied Clinical Medical Physics 24 (10): e14127. doi:10.1002/acm2.14127. |
| Date |
2023 |
| Author |
Kapoor, Rishabh; Sleeman IV, William C.; Ghosh, Preetam; Palta, Jatinder |
| Permission (Reusing this file) |
|
| Other versions |
Licensing
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 00:10, 10 May 2024 |  | 1,367 × 630 (153 KB) | Shawndouglas (talk | contribs) |
You cannot overwrite this file.
File usage
The following page uses this file:
















