Histopathology image classification: Highlighting the gap between manual analysis and AI automation
Contents
| tryptophanase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Tryptophanase tetramer, E.Coli | |||||||||
| Identifiers | |||||||||
| EC no. | 4.1.99.1 | ||||||||
| CAS no. | 9024-00-4 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
The enzyme tryptophanase (EC 4.1.99.1) catalyzes the chemical reaction
- L-tryptophan + H2O indole + pyruvate + NH3
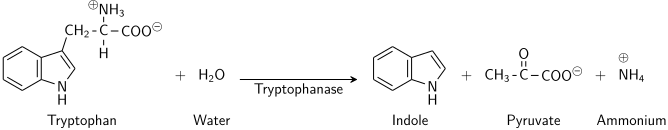
This enzyme belongs to the family of lyases, specifically in the "catch-all" class of carbon-carbon lyases. The systematic name of this enzyme class is L-tryptophan indole-lyase (deaminating; pyruvate-forming). Other names in common use include L-tryptophanase, and L-tryptophan indole-lyase (deaminating). This enzyme participates in tryptophan metabolism and nitrogen metabolism. It has 2 cofactors: pyridoxal phosphate, and potassium.[1][2][3]
Structural studies
As of late 2007, 3 structures have been solved for this class of enzymes, with PDB accession codes 1AX4,[4] 2C44,[5] and 2OQX.[6]
References
- ^ BURNS RO, DEMOSS RD (1962). "Properties of tryptophanase from Escherichia coli". Biochim. Biophys. Acta. 65 (2): 233–44. doi:10.1016/0006-3002(62)91042-9. PMID 14017164.
- ^ Cowell JL, Maser K, DeMoss, RD (1973). "Tryptophanase from Aeromonas liquifaciens. Purification, molecular weight and some chemical, catalytic and immunological properties". Biochimica et Biophysica Acta. 315: 449–463. doi:10.1016/0005-2744(73)90276-3.
- ^ NEWTON WA, MORINO Y, SNELL EE (1965). "Properties of Crystalline Tryptophanase". J. Biol. Chem. 240 (3): 1211–8. doi:10.1016/S0021-9258(18)97562-9. PMID 14284727.
- ^ 1AX4 Retrieved from Protein Data Bank (PDB)
- ^ 2C44 Retrieved from Protein Data Bank (PDB)
- ^ 2OQX Retrieved from Protein Data Bank (PDB)
External links
 Media related to Tryptophanase at Wikimedia Commons
Media related to Tryptophanase at Wikimedia Commons


















