Creating learning health systems and the emerging role of biomedical informatics
| Full article title | Creating learning health systems and the emerging role of biomedical informatics |
|---|---|
| Journal | Learning Health Systems |
| Author(s) | Kohn, Martin S.; Topaloglu, Umit; Kirkendall, Eric S.; Dharod, Ahay; Wells, Brian J.; Gurcan, Metin |
| Primary contact | Email: mkohn at wakehealth dot edu |
| Editors | Wake Forest School of Medicine's Center for Biomedical Informatics |
| Year published | 2022 |
| Volume and issue | 6(1) |
| Article # | e10259 |
| DOI | 10.1002/lrh2.10259 |
| ISSN | 2379-6146 |
| Distribution license | Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International |
| Website | https://onlinelibrary.wiley.com/doi/10.1002/lrh2.10259 |
| Download | https://onlinelibrary.wiley.com/doi/epdf/10.1002/lrh2.10259 (PDF) |
Abstract
Introduction: The nature of information used in medicine has changed. In the past, we were limited to routine clinical data and published clinical trials. Today, we deal with massive, multiple data streams and easy access to new tests, ideas, and capabilities to process them. Whereas in the past getting information for decision-making was a challenge, today's clinicians have readily available access to information and data through the multitude of data-collecting devices, though it remains a challenge at times to analyze, evaluate, and prioritize it. As such, clinicians must become adept with the tools needed to deal with the era of big data, requiring a major change in how we learn to make decisions. Major change is often met with resistance and questions about value. A "learning health system" (LHS) is an enabler to encourage the development of such tools and demonstrate value in improved decision-making.
Methods: In this work, we describe how we are developing a biomedical informatics program to help our medical institution's evolution as an academic LHS, including strategy, training for house staff, and examples of the role of informatics from operations to research.
Results: We highlight an array of LHS implementations and educational programs to improve healthcare and prepare a cadre of physicians with basic information technology (IT) skills. The programs have been well accepted with, for example, increasing interest and enrollment in the educational programs.
Conclusions: We are now in an era when large volumes of a wide variety of data are readily available. The challenge is not so much in the acquisition of data, but in assessing the quality, relevance, and value of the data. The data we can get may not be the data we need. In the past, sources of data were limited, and trial results published in journals were the major source of evidence for decision making. The advent of powerful analytics systems has changed the concept of evidence. Clinicians will have to develop the skills necessary to work in the era of big data. It is not reasonable to expect that all clinicians will also be data scientists. However, understanding the role of artificial intelligence (AI) and predictive analytics, and how to apply them, will become progressively more important. Programs such as the one being implemented at Wake Forest fill that need.
Keywords: biomedical informatics, learning health systems, physician education
Introduction
The nature of data and evidence in healthcare is experiencing rapid change. Personalized healthcare—making decisions that are more likely to benefit the individual—is evolving and requires more kinds of data than has been used until recently. Personalized decision making requires moving beyond the traditional data sources to understand the individual more fully.[1] The techniques used in the past, such as the null hypothesis analysis in randomized controlled studies (RCTs), are still important but insufficient to achieve personalization. The myriad streams of data that may influence health (environmental, domestic, ethnic, cultural, access, political views, wearables, genomics, etc.) leave us in the position of not understanding which of the streams is more important or how they interact. The recent creation of the sub-specialty of clinical informatics confirms that more sophisticated approaches, such as artificial intelligence (AI) and machine learning (ML), are required to use both the usual clinical data and real-world data. Clinicians must become adept with the tools needed to deal with the era of big data, requiring a major change in how we learn to make decisions.[2]
Innovation in health information technology (HIT), such as the electronic health record (EHR), has often been met with resistance and frustration because it was perceived as requiring extensive training, not supporting clinical workflow, requiring additional time to use, or interfering with interactions with the patient, ultimately not providing sufficient value.
The implementation of AI faces similar challenges. Will it show enough value in achieving improved decision making to justify the time and energy necessary to convince stakeholders to use it? Some of the obstacles to adoption include being comfortable with the way things have always been done, having anxiety about learning new skills, and having concerns about the loss of autonomy or the “art” of medicine.
Physicians, after undergoing many years of training, may be particularly resistant to change and the adoption of new technologies. Resistance to the adoption of new HIT often results from a lack of perceived usefulness, perceived threats, and perceived inequities. If physicians believe that a new technology is not useful, especially if it results in more complicated workflow or diminishes their role, it will be difficult to facilitate adoption.
Common human psychological biases may also make it difficult for clinicians to use the information from personalized predictive analytics. This includes confirmation bias (the tendency to emphasize data that confirms one's prior beliefs), the availability heuristic (being more influenced by memorable cases in one's clinical experience), and the endowment effect (the sense of loss from losing what you have, e.g., a cherished belief or conviction).[3]
Overcoming these obstacles requires designs consistent with healthcare's broad workflow, education goals, and demonstration of value. The "learning health system" (LHS) provides the structure to meet those needs. Including concepts of the LHS early in the educational experience is valuable. The Center for Biomedical Informatics at the Wake Forest School of Medicine (WFBMI) is developing a research and educational program to support the inclusion of LHS concepts into mainstream healthcare.
Requirements of a learning health system
In the past, the evidence for decision-making developed slowly and episodically. A topic was studied with a series of RCTs, with the result becoming the standard until the next study was published. The feedback loop to evaluate the impact could be months or years long. Such a lengthy process does not support personalized healthcare. With sophisticated analytics and rapid access to data, we can update the system with ongoing feedback. An LHS embodies such a loop. Data is converted to knowledge, knowledge is used to perform an intervention, the results of the intervention provide new data, and the process cycles continuously.
The elusive LHS is often described by its characteristics[4]:
- Every patient's characteristics and experiences are securely available as data to learn from.
- Practice knowledge derived from these data is immediately available to support health-related decisions by individual members of society, care providers, and managers and planners of health services.
- Improvement is continuous, through ongoing study addressing multiple health-improvement and related goals.
- A socio-technical infrastructure enables this to happen routinely, with a significant level of automation, and with economy of scale.
- Stakeholders within the system view the above activities as part of their culture.
Working effectively in an LHS requires education and training. We are dealing with increasing amounts of data, and more kinds of data than ever before. Identifying the valuable data and discerning patterns that allow better decisions is a new skill. We are moving in that direction, and biomedical informatics, particularly clinical informatics, is key in those efforts. Clinical informatics is the newest clinical sub-specialty, and training programs and fellowships in informatics are becoming common. Creating the environment to support the development of an LHS is an important goal in which the WFBMI is an active participant.
A by-product of the widespread digitization of healthcare over the last two decades is the large amounts of “digital exhaust” (i.e., information collected from the internet by a person or an organization) from technology supporting the clinical and administrative process of delivering care. In fact, as in other systems outside of healthcare, the creation and existence of data is exponentially growing.[5] This data can be leveraged to the benefit of patients, families, providers and staff, as well as the healthcare system at large. One of the main challenges that now exists is how to process and analyze the data to gather actionable insight and knowledge from it. One can easily get lost in “big data” when mining it, hence the need for data scientists and domain experts well-versed in healthcare processes, i.e., clinical informaticians. Without being familiar with the clinical delivery system, the data will not be viewed in the right context to allow this synthesis to occur, and for intervention opportunities to become clear.
Examples of using data to create knowledge to improve practice and operations
Horwitz et al.[6] have published a prime example of how to systematically identify opportunities to apply LHS principles and create value from those activities. In their approach, they largely applied quality improvement and data-driven approaches to achieve favorable outcomes without expending a large number of resources. Much of their value came from “exnovating” or removing unproven activities that required resource spend for what was discovered to be no gain. We are following their lead and beginning our own process, as are many others.
EHRs are tremendous sources of data, and clinical decision support (CDS) in particular is a rich target for much of our work. In 2019 alone, there were 29 million best-practice advisories that were visible to providers and staff, many of which were not acted upon and only served to generate noise in our clinical care delivery processes if they were providing no value. We have begun to systematically and rigorously examine the data for the best opportunities and have already identified large sources of inefficiencies. For example, several of our ambulatory clinics are among the top locations where an influenza vaccination reminder alert has fired, yet several of these clinics do not even have the vaccination stocked and available. Exemption of one clinic alone from receiving the reminder should save about 20,000 alerts from being seen by providers who cannot act on them. Given a conservative estimate of an average alert interrogation time (time a provider spends reading an alert) of five seconds, this equates to about 28 hours of provider time in one flu season, for one clinic site. These alerts are largely targeting physicians and other high-cost staff. A simple configuration change to the alert would save thousands of dollars, and more importantly free up clinician time, in one single clinic.
In the example above, data that are readily available is being used to generate insight about practice patterns, which is then used to create practice efficiencies and optimize provider time. By utilizing data to identify instances where care is not being optimized—but is creating useless, inefficient “busy work”—we are replacing that work with the freedom to do other activities to improve patient care, while improving provider satisfaction and removing a source of ire of physicians.[7]
Developing a biomedical informatics program to help an institution's evolution as a learning health system
When we created the biomedical Informatics program at Wake Forest School of Medicine (WFSoM) in 2018, we designed its structure to help Wake Forest's evolution as an LHS. Wake Forest's vision is to be “a preeminent learning health system that promotes better health for all through collaboration, excellence and innovation.”[8] This cross-disciplinary initiative is designed to integrate resources throughout WFSoM, Wake Forest Baptist Medical Center, and Wake Forest University while complementing the work of other research centers, including the Wake Forest Clinical and Translational Science Institute (CTSI). To achieve this vision, the program pursues three interrelated goals:
- Catalyze: Integrate resources for research informatics across Wake Forest Baptist Medical Center and Wake Forest University, and synergistically support the informatics needs of other research centers, departments, and institutes.
- Discover: Facilitate biomedical discovery across the spectrum of informatics, including bioinformatics, imaging, clinical, translational, and public health informatics, and develop a portfolio of externally funded research in the area of biomedical informatics.
- Educate: Support and inspire the training of the next generation of investigators in the principles and practice of biomedical informatics.
WFBMI's plans for innovation through AI[9] will take many forms, including building and implementing EHR-based clinical alerts, decision support tools, structured data capture, and order sets, as well as facilitating pragmatic clinical trials by being able to randomize by clinics/hospitals. Some driving projects include computer-assisted assessment of otoscopy imaging[10][11][12], grading of follicular lymphoma slides[13][14][15], chest pain risk stratification[16], prediction of glycated hemoglobin values[17], and development of an EHR-based frailty score.[18]
Operational informatics
A well-functioning LHS would need informatics science, as well as the capability of generating knowledge and improving care delivery. WFBMI strives to establish a data infrastructure that enables repeatable and cost-effective learning. Starting with the computational needs of the researchers and business operations at our institution, WFBMI has invested prudently in elastic computing infrastructures resulting from contractual engagements and business associate agreements (BAAs) with the Google Cloud platform (GCP) and Microsoft Azure separately. The GCP serves as the flexible, research computing framework, with available tools to address the needs of precision medicine and other large-scale computing endeavors. WFBMI also utilizes Microsoft Azure as a common computing model for several data sharing and federated learning projects with three other medical centers in the region.
While the reuse of clinical data is hampered by the still prevailing disconnect between the data standards used in patient care and the ones used in clinical research, some efforts to bridge this gap are finally reaping their benefits in the form of clinical research and EHR system integrations.[19]
Substitutable Medical Applications and Reusable Technologies (SMART) on Fast Healthcare Interoperability Resources (FHIR) is a web based standard platform that enables developing solutions to enhance interoperability with EHRs and other systems, including clinical research data capture (e.g., REDCap). We have implemented a SMART on FHIR tool called COMprehensive Post-Acute Stroke Services - Care Plan (COMPASS-CP).[20] COMPASS-CP is a patient-centered application for increased interoperability with any capable EHR. COMPASS-CP captures patient-reported outcome measures (PROMs) of functional and social determinates of health, including priorities from the patient and caregivers at the point of care. The COMPASS-CP algorithms evaluate the data captured in questionnaires and identify factors likely to influence recovery, health, and independence of the stroke survivor across each dimension of care and needed referrals for community-based resources. These are used to generate the patient-facing COMPASS-CP, entitled "Finding My Way Forward for Recovery, Health, and Independence." Care plans provide education, recommendations, and referrals across essential domains of self-management and care, anchored to the four cardinal directions of a compass:
- Numbers: Know your blood pressure, hemoglobin A1c, cholesterol, etc.
- Engage: Be active in mind, body, and spirit through physical, cognitive, and social activity.
- Support: Seek support for your stress, family stress, finances for medications, and transportation.
- Willingness: Be willing to manage your medications and lifestyle.
Results and details of the 40 hospitals and 8,000 patients who participated in the COMPASS-CP pragmatic trial can be found in Duncan et al.[20]
Another example of the use of FHIR technology is the Mobile Patient Technology for Health (mPATH) technology, designed to enhance patient involvement in decisions for colorectal cancer screening.
Traditionally, decision aids have been provider-focused, but they are increasingly becoming more patient-centric. Miller et al. developed the mPATH tablet computer-based tool, that provides educational material regarding colorectal cancer screening options.[21] After rooming, patients are given a tablet computer that provides educational material about the pros and cons of different screening options. While patients have the opportunity to discuss the options with the clinician during the visit, ultimately the patient “self-orders” the preferred screening test (e.g., fecal blood test or colonoscopy). Patients who self-order a colorectal cancer screening test receive text messages to remind them about the test, to provide encouragement, and to ask if patients have questions. Subsequent text messages are sent to patients in order to encourage completion of the screening test. A randomized controlled trial demonstrated that screening was twice as likely to be completed (30%) in the intervention group as compared to the control group (15%).
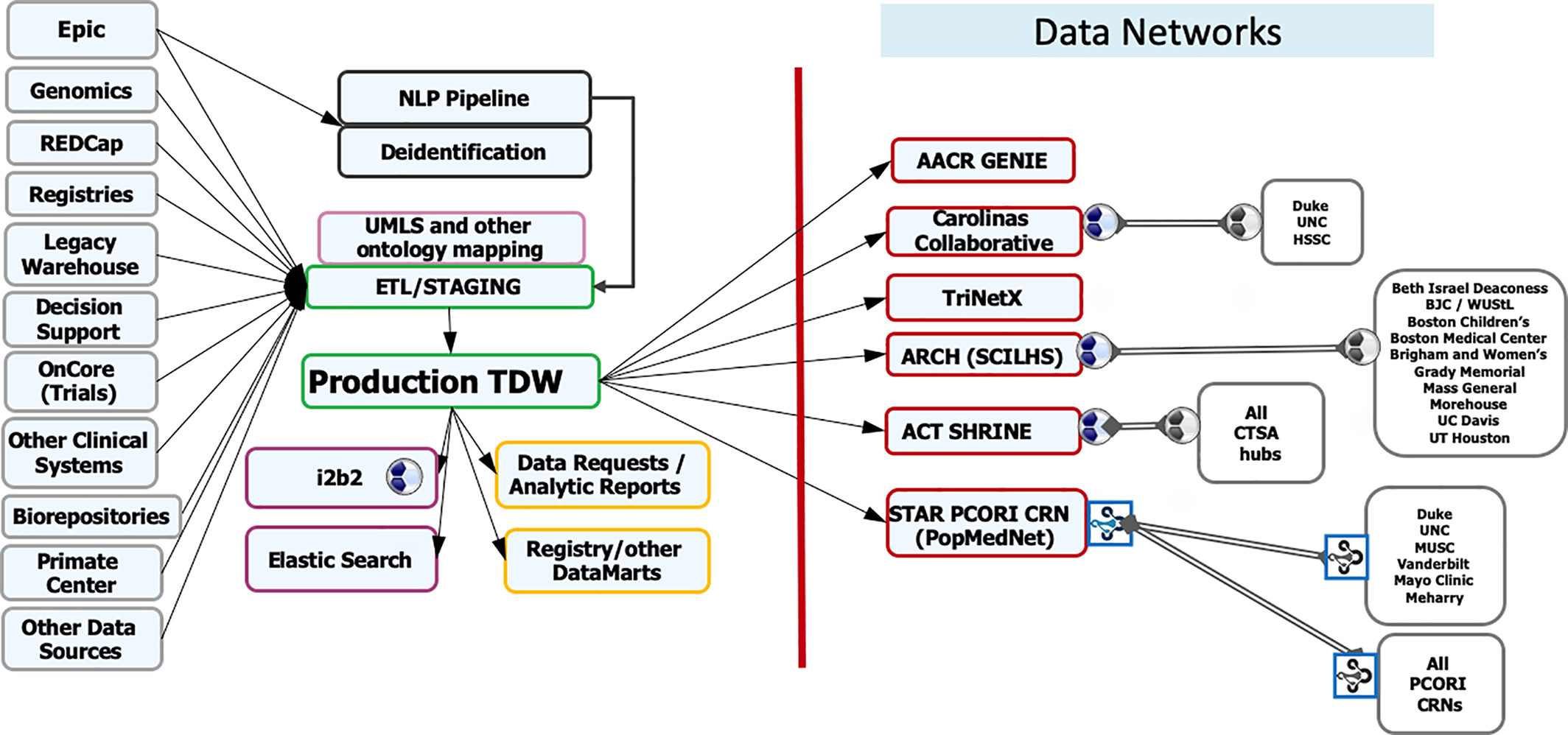
The design of this project was built on research showing that multi-level CDS tools are more likely to be effective.[21] For example, Roshanov showed that CDS systems were much more likely to succeed when the tools provided advice for patients in addition to practitioners.[22] The study design should be given significant credit for the success of this project, and it highlights the need for informatics professionals who are familiar with the literature to be involved in the implementation of new informatics tools. The WFBMI has built the Translational Data Warehouse (TDW) to facilitate a comprehensive and semantically rich data ecosystem. The development of our translational data warehouse preceded the formation of the WFBMI. The TDW serves as a central resource for a gamut of research and quality improvement needs in the institution (Figure 1). Data is mapped to the standards, including RxNorm, LOINC, NAACR, etc., and extracted from primary sources via the use of extract, transform, load (ETL). Mapping to standard terminologies in the TDW provides a consistent means for researchers to collaborate with other institutions, and facilitates participation in several clinical data research networks (CDRNs) by conforming local data to these networks' Common Data Models (CDMs).
We are in the planning phase of utilizing Epic-supported FHIR resources as an additional mechanism for data transfer to the TDW. WFBMI is also committed to developing a semantic framework that will minimize data misinterpretation and discovery challenges. We maintain a terminology server to house Unified Medical Language System (UMLS) standard terminologies. It serves vocabularies and “ontologies” for multiple research processes for a cohesive interoperability SNOMED Clinical Terms (SNOMED CT), LOINC, and NCI Common Data Elements (CDEs). In addition, our NCI-trained staff creates CDEs in the Cancer Data Standards Registry and Repository (caDSR), which are used in case report forms (CRFs), as required for all cancer studies. To facilitate reuse, we currently use existing CRFs from our CRF library or create new CRFs that have been curated with CDEs.
|
In addition to staff-facilitated data requests, investigators can perform queries on the TDW using i2b2 and, upon appropriate IRB approval, they can download automatically generated datasets through a self-service tool. This internally developed tool interfaces directly with the IRB system to determine authorizations and can deliver Excel exports of data in minutes, greatly reducing turnaround times (TAT) on data retrievals. In addition to EHR data, the TDW has been linked to outside data sources, most notably U.S. census data and the North Carolina State Death Registry. TDW is also our conduit for participation in the federated data networks for obtaining larger sample sizes (particularly important for rarer disease investigations) and realization of the full potential of population research. WFBMI enables WFSoM's involvement and contributes to regional and national CDRNs, including Carolina Collaborative, STAR (Mid-South) PCORnet Clinical Data Research Network, Accruals to Clinical Trials (ACT) Network of NCATS, TriNetX, and AACR Genomics Evidence Neoplasia Information Exchange (GENIE). Participation in these networks has led to dozens of clinical trial activations and funded collaborative studies.
Education and training in a learning health system
The LHS is a new paradigm for the academic medical center requiring a more diverse skill set for faculty to be successful in clinical care, quality improvement, and research rather than what was needed in the past. Many of the skills necessary for a well-functioning LHS already exist but are scattered across multiple disciplines, making interdisciplinary teams very important. A project that involves the extraction and analyses of clinical data to create knowledge and tools for implementation into clinical care requires, at a minimum, knowledge of health information systems, relational databases, statistics, clinical care, and implementation science. The field of clinical informatics rests at the intersection of these different domains and has been a focus of our educational efforts in the CTSI. We have targeted junior faculty in both clinical and non-clinical departments for participation in an annual Clinical Informatics Short Course for the past four years. The course consists of 16 hours of in-person classroom instruction separated into two-hour or four-hour blocks. Course evaluations by physicians showed that ~75% of students felt the course met their educational needs, ~85% reported that the course increased their competence in this area, and ~85% said the course would improve their patient outcomes. Although the student evaluations have been positive, some evaluations indicated that the course did a good job of highlighting the difficulties in clinical informatics but did not offer specific skills for solutions. In response to this feedback, the format of the course has evolved from a primarily didactic and discussion-based curriculum to a more experiential approach.
The 2019 class worked together to extract and analyze data to create a model for predicting pneumonia readmission risk, which was presented to the Society for Medical Decision Making.[23] The 2020 class featured 26 student enrollees and involved hands-on machine learning with the R programming language to develop risk prediction models from data extracted from the EHR. The course was shifted from an in-person to a virtual format via WebEx due to the COVID-19 pandemic, but the majority of students who completed the course still rated it well overall and would recommend the course to a colleague. One student even expressed interest in re-taking the course in the future. Some of the students reflected that they had insufficient R programming skills to receive maximum benefit from the course. The course is synchronized to occur immediately following a preexisting R beginner course for those students who need the additional training. The WFBMI faculty have also developed a Master of Science level course entitled “Introduction to Biomedical Informatics” that has been offered to our Translational and Health System Science graduate students since 2019. The overview course focuses heavily on health informatics and includes topics on medical decision making, clinical decision support, cloud-based computing, image analyses, and common data models. The 2020 course received overwhelming interest and needed to be capped at ~30 students.
Other educational initiatives
Clinical Informatics Board Certification Subsidy: Physicians wishing to become board certified in clinical informatics may have the opportunity to sit for the board examination through the “practice-pathway” without completing a formal two-year clinical informatics fellowship. We discovered that a significant number of clinical faculty have participated in clinical informatics-related activities that make them eligible. The $2,000 subsidies help to offset the cost of a board review course offered by the American Medical Informatics Association (AMIA) and the examination fee. We plan to offer the subsidies until the practice pathway closes after the 2022 examination.
EHR mini-course : The EHR-mini course offers a two-hour overview of the opportunities and complexities involved with the secondary use of clinical data for research. A second two-hour session includes discussion of project ideas generated by the class and provides more details about the specific resources available through the CTSI and the logistics for completing a project. We have begun offering “mini-pilot” awards of up to $3,000 that are designed to provide enough funding to push an idea forward with the hope that the results could lead to the development of a larger pilot application.
Titles of the three “mini-pilots” awarded in 2019 were:
- Risk prediction modeling of missed appointments in pediatric subspecialty clinics
- Diagnosis and Management of ARDS in a Tertiary Care Pediatric Intensive Care Unit
- Patient- and Provider- Reported Outcomes, Disease Features, and Medication Exposure in Children with Juvenile Idiopathic Arthritis
'EHR vendor-based certification: The CTSI will begin providing funding for physician-researchers to become certified through our EHR vendor to build optimization tools. This training will make these investigators more aware of the EHR capabilities to support both clinical care and research. It is hoped that this knowledge will facilitate LHS research that sits on the boundary of quality improvement and clinical care. We are working with IT in order to integrate the trainees with existing projects to provide hands-on experience after certification. IT will benefit from the clinical knowledge being provided to this collaboration.
The Wake Forest Clinical Scholars in Informatics (CSI) program
The LHS requires a constant feedback loop with input from a learning community. It is essential to train physicians to be competent in the fundamental components of biomedical informatics, data science, and applied clinical informatics. In an academic learning health system, resident physicians are often the closest individuals to the “ground floor” and understand the challenges in health services delivery and clinical care. We hypothesize there is value for an applied clinical informatics pathway for resident physicians as a distinct and separate entity of the clinical informatics fellowship. Many physicians will have an interest and require basic knowledge and skills regarding applied clinical informatics but not in pursuing a full clinical informatics board certification and/or fellowship. Having these skills independent of specialty is important to further our goals as a learning healthcare system.
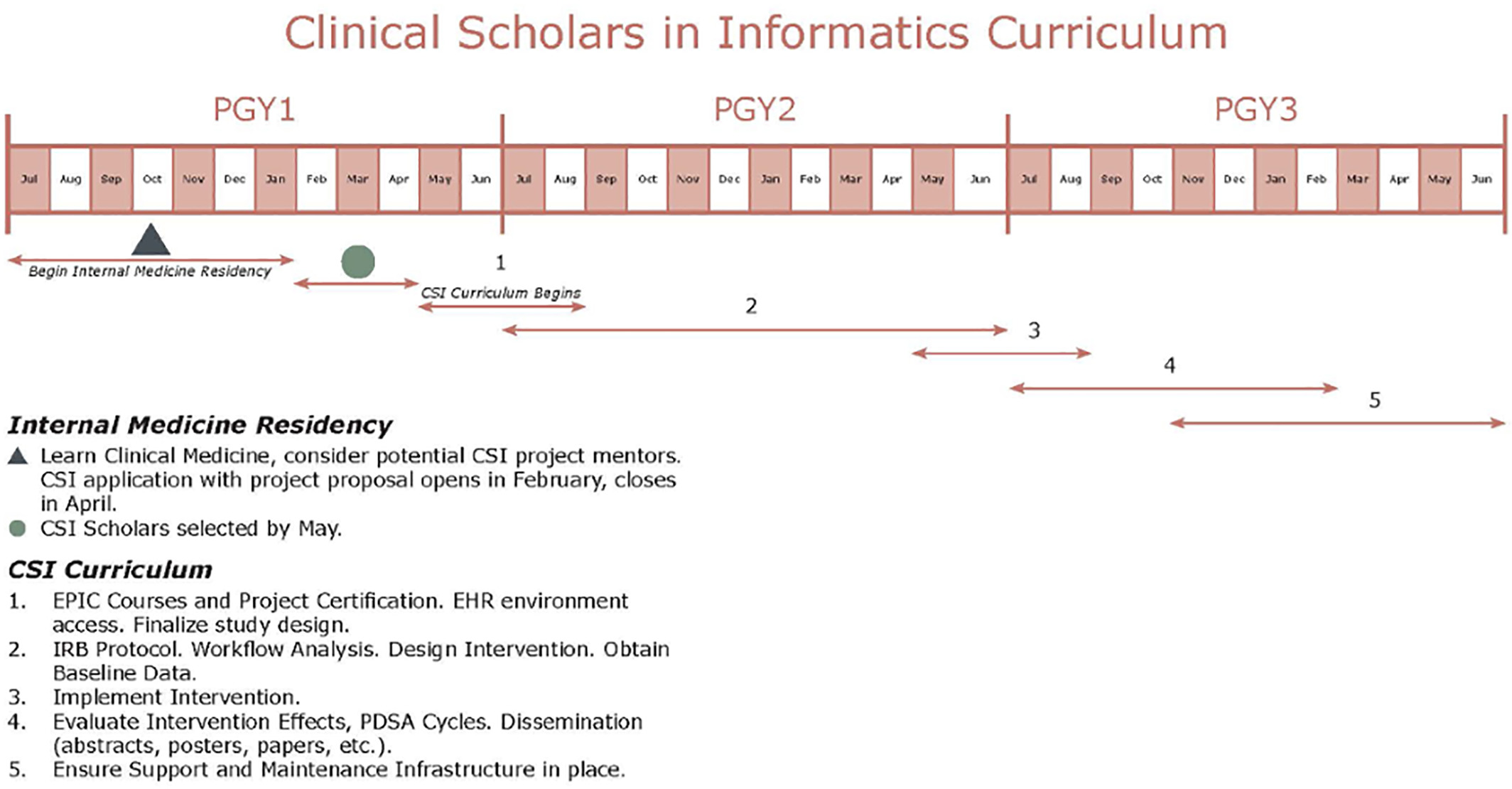
The Clinical Scholars in Informatics (CSI) program is a low-cost applied clinical informatics educational and mentorship pathway by way of the design and implementation of an applied informatics project. In the current form, CSI is a two-year program designed for internal medicine (IM) residents. Two PGY first-year residents are selected for the pathway to begin in their PGY second year and complete upon graduation from IM residency. CSI will develop a cadre of physicians with basic health IT competency. We perceive CSI as operationalization of the LHS designed for resident trainees. CSI residents are expected to design and implement an informatics-related intervention leveraging the EHR by the end of their PGY second year. Furthermore, CSI residents are expected to evaluate the impact of their intervention within the health system. Finally, CSI residents are expected to present their work at local, regional, and national conferences and submit a manuscript in a relevant journal (Figure 2).
|
The CSI is different from a traditional research program with a stipend. CSI residents receive EHR certification with special privileges to modify the EHR in collaboration with Information Technology Services. The CSI focuses on designing and implementing an intervention to impact direct patient care. These interventions are often pain points in clinical workflows or evidence that exists in the literature with intermittent or poor adherence in the real world. Furthermore, CSI residents are expected to participate in a monthly journal club that specifically addresses educational informatics topics aligned with the progress that the CSI resident should be making with respect to their applied informatics project. Example journal club topics include workflow analysis, clinical decision support, data standards, change management, basic coding concepts, regulatory issues (HIPAA, HITECH, TJC, etc.), security, and privacy. The curriculum is noted in Figure 2. A substantial focus on support and maintenance of the interventions is considered early in the process, an often overlooked component of many projects.
The overall cost of the CSI program is approximately $5,000 per scholar per year, which includes EHR building certification, stipend, and a laptop. Notably, personnel costs (faculty time, support from EHR analysts and data analysts) are not included in the $5,000 per scholar per year cost. Each CSI scholar is paired with an informatics mentor, a specialty mentor, an EHR analyst, and a data analyst to ensure all components of the applied informatics intervention, including EHR build/configuration and EHR data extraction and visualization, can be accomplished. A comprehensive list of peer-reviewed posters, presentations, manuscripts, and grants can be found on the CSI website.[24]
Conclusions
The CSI is a low-cost, novel applied clinical informatics resident pathway, which can be broadly applied to most internal medicine programs across the United States. There are clearly bidirectional net benefits for the resident physicians and for the health system as a whole. The residents benefit through the experience of driving an applied informatics project end-to-end and deriving academic capital. The health system benefits by implementation of a locally relevant applied clinical informatics project with robust evaluation. Additionally, the CSI program functions as internal development and recruitment for broad-based informatics expertise across the enterprise. Future directions include scaling to additional departments, formalizing the curriculum with Wake Forest graduate medical education, and defining a sustainable funding stream (institutional vs extramural).[24]
The WFBMI is engaged in a multi-faceted program to improve clinical-decision making and health care operations by preparing for the data-driven future. The program emphasizes data acquisition, management and analysis, as well as training clinicians to thrive in the big data era. Value-based healthcare is dependent on such programs.
We are now in an era when large volumes of a wide variety of data are readily available. The challenge is not so much in the acquisition of data, but in assessing the quality, relevance, and value of the data. The data we can get may not be the data we need. In the past, sources of data were limited, and trial results published in journals were the major source of evidence for decision-making. The advent of powerful analytics systems has changed the concept of evidence. Clinicians will have to develop the skills necessary to work in the era of big data. It is not reasonable to expect that all clinicians will also be data scientists. However, understanding the role of AI and predictive analytics, and how to apply them, will become progressively more important. Programs such as the one being implemented at Wake Forest fill that need.
Abbreviations
- ACT: Accruals to Clinical Trials
- AI: artificial intelligence
- AMIA: American Medical Informatics Association
- BAA: business associate agreement
- caDSR: Cancer Data Standards Registry and Repository
- CDE: Common Data Element
- CDM: Common Data Model
- CDRN: clinical data research network
- CDS: clinical decision support
- COMPASS-CP: COMprehensive Post-Acute Stroke Services - Care Plan
- CRF: case report form
- CSI: Clinical Scholars in Informatics
- CTSI: Wake Forest Clinical and Translational Science Institute
- EHR: electronic health record
- ETL: extract, transform, load
- FHIR: Fast Healthcare Interoperability Resources
- GCP: Google Cloud platform
- GENIE: Genomics Evidence Neoplasia Information Exchange
- HIT: health information technology
- IM: internal medicine
- IT: information technology
- LHS: learning health system
- ML: machine learning
- mPATH: Mobile Patient Technology for Health
- PROM: patient-reported outcome measure
- RCT: randomized controlled study
- SMART: Substitutable Medical Applications and Reusable Technologies
- TAT: turnaround time
- TDW: Translational Data Warehouse
- UMLS: Unified Medical Language System
- WFBMI: Center for Biomedical Informatics at the Wake Forest School of Medicine
- WFSoM: Wake Forest School of Medicine
Acknowledgements
Conflict of interest
The authors have no conflict of interest to report.
References
- ↑ Pencina, Michael J.; Peterson, Eric D. (26 April 2016). "Moving From Clinical Trials to Precision Medicine: The Role for Predictive Modeling". JAMA 315 (16): 1713–1714. doi:10.1001/jama.2016.4839. ISSN 1538-3598. PMID 27115375. https://pubmed.ncbi.nlm.nih.gov/27115375.
- ↑ Krumholz, Harlan M. (1 July 2014). "Big Data And New Knowledge In Medicine: The Thinking, Training, And Tools Needed For A Learning Health System". Health affairs (Project Hope) 33 (7): 1163–1170. doi:10.1377/hlthaff.2014.0053. ISSN 0278-2715. PMC 5459394. PMID 25006142. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5459394/.
- ↑ Ubel, Peter A.; Asch, David A. (1 February 2015). "Creating value in health by understanding and overcoming resistance to de-innovation". Health Affairs (Project Hope) 34 (2): 239–244. doi:10.1377/hlthaff.2014.0983. ISSN 1544-5208. PMID 25646103. https://pubmed.ncbi.nlm.nih.gov/25646103.
- ↑ Friedman, Charles P.; Allee, Nancy J.; Delaney, Brendan C.; Flynn, Allen J.; Silverstein, Jonathan C.; Sullivan, Kevin; Young, Kathleen A. (29 November 2016). "The science of Learning Health Systems: Foundations for a new journal". Learning Health Systems 1 (1): e10020. doi:10.1002/lrh2.10020. ISSN 2379-6146. PMC 6516721. PMID 31245555. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6516721/.
- ↑ Dinov, Ivo D. (2016). "Volume and Value of Big Healthcare Data". Journal of medical statistics and informatics 4: 1–7. doi:10.7243/2053-7662-4-3. ISSN 2053-7662. PMC 4795481. PMID 26998309. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4795481/.
- ↑ Horwitz, Leora I.; Kuznetsova, Masha; Jones, Simon A. (19 September 2019). "Creating a Learning Health System through Rapid-Cycle, Randomized Testing". The New England Journal of Medicine 381 (12): 1175–1179. doi:10.1056/NEJMsb1900856. ISSN 1533-4406. PMID 31532967. https://pubmed.ncbi.nlm.nih.gov/31532967.
- ↑ Marmor, Rebecca A.; Clay, Brian; Millen, Marlene; Savides, Thomas J.; Longhurst, Christopher A. (1 January 2018). "The Impact of Physician EHR Usage on Patient Satisfaction". Applied Clinical Informatics 9 (1): 11–14. doi:10.1055/s-0037-1620263. ISSN 1869-0327. PMC 5801886. PMID 29298451. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5801886/.
- ↑ "About the School". Wake Forest School of Medicine. 2021. https://school.wakehealth.edu/About-the-School.
- ↑ Niazi, Muhammad Khalid Khan; Parwani, Anil V.; Gurcan, Metin N. (1 May 2019). "Digital pathology and artificial intelligence". The Lancet. Oncology 20 (5): e253–e261. doi:10.1016/S1470-2045(19)30154-8. ISSN 1474-5488. PMC 8711251. PMID 31044723. https://pubmed.ncbi.nlm.nih.gov/31044723.
- ↑ Moberly, Aaron C.; Zhang, Margaret; Yu, Lianbo; Gurcan, Metin; Senaras, Caglar; Teknos, Theodoros N.; Elmaraghy, Charles A.; Taj-Schaal, Nazhat et al. (1 August 2018). "Digital otoscopy versus microscopy: How correct and confident are ear experts in their diagnoses?". Journal of Telemedicine and Telecare 24 (7): 453–459. doi:10.1177/1357633X17708531. ISSN 1758-1109. PMID 28480781. https://pubmed.ncbi.nlm.nih.gov/28480781.
- ↑ Senaras, Caglar; Moberly, Aaron C.; Teknos, Theodoros; Essig, Garth; Elmaraghy, Charles; Taj-Schaal, Nazhat; Yu, Lianbo; Gurcan, Metin (3 March 2017). Armato, Samuel G.; Petrick, Nicholas A.. eds. Autoscope: automated otoscopy image analysis to diagnose ear pathology and use of clinically motivated eardrum features. Orlando, Florida, United States. pp. 101341X. doi:10.1117/12.2250592. http://proceedings.spiedigitallibrary.org/proceeding.aspx?doi=10.1117/12.2250592.
- ↑ Senaras, Caglar; Moberly, Aaron C.; Teknos, Theodoros; Essig, Garth; Elmaraghy, Charles; Taj-Schaal, Nazhat; Yua, Lianbo; Gurcan, Metin N. (27 February 2018). Mori, Kensaku; Petrick, Nicholas. eds. "Detection of eardrum abnormalities using ensemble deep learning approaches". Medical Imaging 2018: Computer-Aided Diagnosis (Houston, United States: SPIE): 45. doi:10.1117/12.2293297. ISBN 978-1-5106-1639-4. https://www.spiedigitallibrary.org/conference-proceedings-of-spie/10575/2293297/Detection-of-eardrum-abnormalities-using-ensemble-deep-learning-approaches/10.1117/12.2293297.full.
- ↑ Senaras, Caglar; Niazi, Muhammad Khalid Khan; Arole, Vidya; Chen, Weijie; Sahiner, Berkman; Shana’ah, Arwa; Louissaint, Abner; Hasserjian, Robert Paul et al. (18 March 2019). Tomaszewski, John E.; Ward, Aaron D.. eds. "Segmentation of follicles from CD8-stained slides of follicular lymphoma using deep learning". Medical Imaging 2019: Digital Pathology (San Diego, United States: SPIE): 25. doi:10.1117/12.2512262. ISBN 978-1-5106-2559-4. https://www.spiedigitallibrary.org/conference-proceedings-of-spie/10956/2512262/Segmentation-of-follicles-from-CD8-stained-slides-of-follicular-lymphoma/10.1117/12.2512262.full.
- ↑ Gurcan, Metin; Senaras, Caglar; Pennell, Michael; Chen, Weijie; Sahiner, Berkman; Shana’ah, Arwa; Louissaint, Abner; Hasserjian, Robert Paul et al. (6 March 2018). Gurcan, Metin N.; Tomaszewski, John E.. eds. "Tumor microenvironment for follicular lymphoma: structural analysis for outcome prediction". Medical Imaging 2018: Digital Pathology (Houston, United States: SPIE): 2. doi:10.1117/12.2295284. ISBN 978-1-5106-1651-6. https://www.spiedigitallibrary.org/conference-proceedings-of-spie/10581/2295284/Tumor-microenvironment-for-follicular-lymphoma--structural-analysis-for-outcome/10.1117/12.2295284.full.
- ↑ Fauzi, Mohammad Faizal Ahmad; Pennell, Michael; Sahiner, Berkman; Chen, Weijie; Shana’ah, Arwa; Hemminger, Jessica; Gru, Alejandro; Kurt, Habibe et al. (30 December 2015). "Classification of follicular lymphoma: the effect of computer aid on pathologists grading". BMC Medical Informatics and Decision Making 15. doi:10.1186/s12911-015-0235-6. ISSN 1472-6947. PMC 4696238. PMID 26715518. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4696238/.
- ↑ Mahler, Simon A.; Lenoir, Kristin M.; Wells, Brian J.; Burke, Gregory L.; Duncan, Pamela W.; Case, L. Douglas; Herrington, David M.; Diaz-Garelli, Jose-Franck et al. (27 November 2018). "Safely Identifying Emergency Department Patients with Acute Chest Pain for Early Discharge: The HEART Pathway Accelerated Diagnostic Protocol". Circulation 138 (22): 2456–2468. doi:10.1161/CIRCULATIONAHA.118.036528. ISSN 0009-7322. PMC 6309794. PMID 30571347. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6309794/.
- ↑ Wells, Brian J; Lenoir, Kristin M; Diaz-Garelli, Jose-Franck; Futrell, Wendell; Lockerman, Elizabeth; Pantalone, Kevin M; Kattan, Michael W (22 October 2018). "Predicting Current Glycated Hemoglobin Values in Adults: Development of an Algorithm From the Electronic Health Record". JMIR Medical Informatics 6 (4). doi:10.2196/10780. ISSN 2291-9694. PMC 6231807. PMID 30348631. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6231807/.
- ↑ Pajewski, Nicholas M; Lenoir, Kristin; Wells, Brian J; Williamson, Jeff D; Callahan, Kathryn E (1 October 2019). "Frailty Screening Using the Electronic Health Record Within a Medicare Accountable Care Organization". The Journals of Gerontology Series A: Biological Sciences and Medical Sciences 74 (11): 1771–1777. doi:10.1093/gerona/glz017. ISSN 1079-5006. PMC 6777083. PMID 30668637. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6777083/.
- ↑ Topaloglu, U.; Niedner, R.H. (2016). "Advances in Clinical Research Toward the Data-Driven Economy" (PDF). Clinical Researcher 30: 58–62. doi:10.14524/CR-16-0008. https://acrpnet.org/wp-content/uploads/dlm_uploads/2017/03/ACRP-Clinical-Researcher-August-2016-1.pdf.
- ↑ 20.0 20.1 Duncan, Pamela W.; Abbott, Rica M.; Rushing, Scott; Johnson, Anna M.; Condon, Christina N.; Lycan, Sarah L.; Lutz, Barbara J.; Cummings, Doyle M. et al. (1 August 2018). "COMPASS-CP: An Electronic Application to Capture Patient-Reported Outcomes to Develop Actionable Stroke and Transient Ischemic Attack Care Plans". Circulation. Cardiovascular Quality and Outcomes 11 (8): e004444. doi:10.1161/CIRCOUTCOMES.117.004444. ISSN 1941-7705. PMID 30354371. https://pubmed.ncbi.nlm.nih.gov/30354371.
- ↑ 21.0 21.1 Miller, David P.; Denizard-Thompson, Nancy; Weaver, Kathryn E.; Case, L. Doug; Troyer, Jennifer L.; Spangler, John G.; Lawler, Donna; Pignone, Michael P. (17 April 2018). "Effect of a digital health intervention on receipt of colorectal cancer screening in vulnerable patients: a randomized, controlled trial". Annals of internal medicine 168 (8): 550–557. doi:10.7326/M17-2315. ISSN 0003-4819. PMC 6033519. PMID 29532054. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6033519/.
- ↑ Roshanov, P. S.; Fernandes, N.; Wilczynski, J. M.; Hemens, B. J.; You, J. J.; Handler, S. M.; Nieuwlaat, R.; Souza, N. M. et al. (14 February 2013). "Features of effective computerised clinical decision support systems: meta-regression of 162 randomised trials" (in en). BMJ 346 (feb14 1): f657–f657. doi:10.1136/bmj.f657. ISSN 1756-1833. https://www.bmj.com/lookup/doi/10.1136/bmj.f657.
- ↑ Lenoir, K.; Barnes, E.; McCrory, M. et al. (2020). "PS 4-58 The Application of a 30-Day Pneumonia Readmission Model at an External Healthcare Institution, 41st Annual Meeting of the Society for Medical Decision Making, Portland, Oregon, October 20–23, 2019" (in en). Medical Decision Making 40 (1). doi:10.1177/0272989X19890544. ISSN 0272-989X. http://journals.sagepub.com/doi/10.1177/0272989X19890544.
- ↑ 24.0 24.1 "Internal Medicine Residency Clinical Scholars in Informatics Pathway". Wake Forest School of Medicine. 2019. https://school.wakehealth.edu/Education-and-Training/Residencies-and-Fellowships/Internal-Medicine-Residency/Curriculum-Overview/Clinical-Scholars-in-Informatics-Pathway.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation, spelling, and grammar. In some cases important information was missing from the references, and that information was added. Several bits of text were inadvertently repeated, and those repetitions were removed. No citation was given for the Wake Forest quote, so one was added. Everything else remains true to the original article, per the "NoDerivatives" portion of the distribution license.