ChromaWizard: An open-source image analysis software for multicolor fluorescence in situ hybridization analysis
| Full article title | ChromaWizard: An open-source image analysis software for multicolor fluorescence in situ hybridization analysis |
|---|---|
| Journal | Cytometry Part A |
| Author(s) | Auer, Norbert; Hrdina, Astrid; Hiremath, Chaitra; Vcelar, Sabine; Baumann, Martina; Borth, Nicole; Jadhav, Vaibhav |
| Author affiliation(s) | Austrian Centre for Industrial Biotechnology |
| Primary contact | Email: nicole dot borth at boku dot ac dot at |
| Year published | 2018 |
| Volume and issue | 93(7) |
| Page(s) | 749–54 |
| DOI | 10.1002/cyto.a.23505 |
| ISSN | 1552-4930 |
| Distribution license | Creative Commons Attribution-NonCommercial 4.0 International |
| Website | https://onlinelibrary.wiley.com/doi/full/10.1002/cyto.a.23505 |
| Download | https://onlinelibrary.wiley.com/doi/epdf/10.1002/cyto.a.23505 (PDF) |
Abstract
Multicolor image analysis finds its applications in a broad range of biological studies. Specifically, multiplex fluorescence in situ hybridization (M‐FISH) for chromosome painting facilitates the analysis of individual chromosomes in complex metaphase spreads and is widely used to detect both numerical and structural aberrations. While this is well established for human and mouse karyotypes, for which species sophisticated software and analysis tools are available, other organisms and species are less well served. Commercially available software is proprietary and not easily adaptable to other karyotypes. Therefore, a publicly available open-source software that combines flexibility and customizable functionalities is needed. Here we present such a tool, called “ChromaWizard,” which is based on popular scientific image analysis libraries (OpenCV, scikit‐image, and NumPy). We demonstrate its functionality on the example of primary Chinese hamster (Cricetulus griseus) fibroblasts metaphase spreads and on Chinese hamster ovary cell lines, known for their large number of chromosomal rearrangements. The application can be easily adapted to any kind of available labeling kits and is independent of the organism and instrumentation used. It allows direct inspection of the original hybridization signals and enables either manual or automatic assignment of colors, making it a functional and versatile tool that can also be used for other multicolor applications.
Keywords: fluorescence in situ hybridization (FISH), chromosome painting, open-source image analysis software
Introduction
fluorescence in situ hybridization (FISH) has greatly facilitated the characterization of gene position, chromosomes, and genome organization.[1] FISH involves the generation and hybridization of loci-specific fluorescence‐labeled nucleic acid probes on to metaphase chromosomes or onto interphase nuclei.[2] The use of this method made gene mapping possible at much higher resolution compared with other physical mapping technologies.[3] FISH was further developed into chromosome painting to simultaneously and unequivocally distinguish chromosomes and their rearrangements in complex karyotypes. Using chromosome-specific probes and combining multiple fluorophores allows the visualization of all chromosomes with individual and unique color combinations.[4] Chromosome painting has proven to be very important not only in diagnostics and in cancer research, but also in biological research for numerical and structural aberrations, as analysis can be done at the entire genome level.[5][6] There have been considerable technical improvements for analysis of human and murine samples. However, even though chromosome painting probes become available for a larger number of non‐model species from commercial or specialized labs[7], this method is still not widely applied due to the absence of open‐source software for analysis.[8]
Specialized cytogenetics labs are equipped with the necessary infrastructure, including highly sophisticated software and instruments specifically set up for the purpose of karyotype analysis and chromosome painting, including examples such as Isis (by Metasystems), CytoVision (by Leica Biosystems), and Hyperspectral (by Applied Spectral Imaging). However, if this method needs to be established and used as a side project in a lab not thus equipped, this might turn into a challenge; while imaging facilities are typically accessible, software packages come at a cost that may not be budgeted for given the restricted budgets of most core facilities.
Only to a limited extent can laboratories turn to free (or open‐source) tools such as ImageJ[9] for general applications, as they are technically challenging and require programming skills to write macros to customize the user application, which again limits their use. Therefore, we introduce a stand‐alone, simple open‐source program called ChromaWizard for analysis and visualization of multicolor fluorescence images that addresses the needs of custom non‐model organism samples. This software integrates image processing, multicolor integration, chromosome separation, and visualization with false color assignments. It processes images for any number of color channels/probes with flexible configuration. One of the important features of ChromaWizard is the use of a GUI to access full functionality so that the user can refine results. The software works with standard image formats (TIFF, PNG, and JPEG) taken either by epifluorescence or confocal microscopes. This open‐source tool is filling a gap for users lacking the ability or time to write macro programs, while being freely accessible for advanced users for further customization to enable specific or enhanced imaging applications. It can be connected with other image processing tools to improve the quality of final images as suggested.[10][11]
In this article we present an overview of the functionality of the program, as well as more detailed descriptions of selected features using chromosome painting analysis performed on primary fibroblasts from the Chinese hamster and on Chinese hamster ovary (CHO‐K1) cells as examples.
Materials and methods
Cell lines and sample preparation
Chinese hamster lung fibroblast cells[12] were maintained in DMEM medium containing 10% of FCS and 2 mM Glutamine, and the cells were incubated at 37°C in 7% CO2.
Suspension and protein-free adapted CHO‐K1 (ECACC CCL‐61) cells were grown in CD‐CHO (Gibco/Life Technologies) supplemented with 8 mM L‐Glu and Anti‐Clumping Agent (Gibco/Life Technologies) in flasks shaken at 140 rpm at 37°C in 7% CO2. For the generation of metaphase spreads, 10 ml of exponentially growing cells were treated with 25 μl of Colcemid stock (100 μg/ml in PBS, Sigma‐Aldrich, USA) to a final concentration of 0.25 μg/ml for up to 16 hours. Mitotic cells were spun down and 5 ml of a 75 M hypotonic potassium chloride solution were added under continuous soft agitation, followed by incubation at 37°C for 20 minutes. Cells were fixed by the addition of 500 μl of a 3:1 cold methanol/acetic acid mixture. The fixation was repeated four times and centrifuged after each addition. Metaphase cells were stored at −80°C in 1 ml fixative.
Chromosome painting
Painting of metaphase chromosome spreads was performed using the 12XCHamster mFISH probe kit (MetaSystems, Germany) according to the vendor's protocol. In this kit, probes for each of the 12 Chinese hamster chromosomes are labeled by a unique combination of five fluorophores (FITC = green; Spectrum Orange = orange; TexasRed = red; DEAC (Diethylamino‐coumarin) = aqua; and Cy5 = NIR). As modification, the incubation period for hybridization was increased from 24 to 48 hours, which provided the best quality images. No DAPI counterstaining was used. After hybridization and washing of the slide, 20 μl antifade mounting reagent (SlowFade Gold Antifade, Thermo Fisher Scientific, Waltham, MA, USA) was applied onto the spread, which was carefully overlaid with a 76 × 26 mm2 coverslip and sealed with transparent nail varnish.
Images of the fluorochrome‐labeled chromosome spreads were recorded with a Leica SP8‐gSTED super‐resolution microscope (Leica Microsystems, Germany) using the following excitation and emission filters: NIR (ex: 644 nm, em: 655–697 nm); RED (ex: 595, em: 625–655 nm); ORANGE (ex: 552 nm, em: 580–611 nm); GREEN (ex: 505 nm, em: 537‐570 nm); AQUA (ex: 405 nm, em: 441–498 nm); mask layer (ex: 518, 551, and 581 nm, em: 600–744 nm). All pictures were taken with 100× magnification (objective: HC PL APO CS2 100×/1.40 OIL) at a scan speed of 400 Hz. The images from the six different channels were recorded sequentially from higher to lower excitation wavelength to reduce crosstalk (Table 1). Images were exported in the TIFF format with LAS X 2.0.1.14392 software (Leica Microsystems, Germany) for further analysis.
Results
Software description
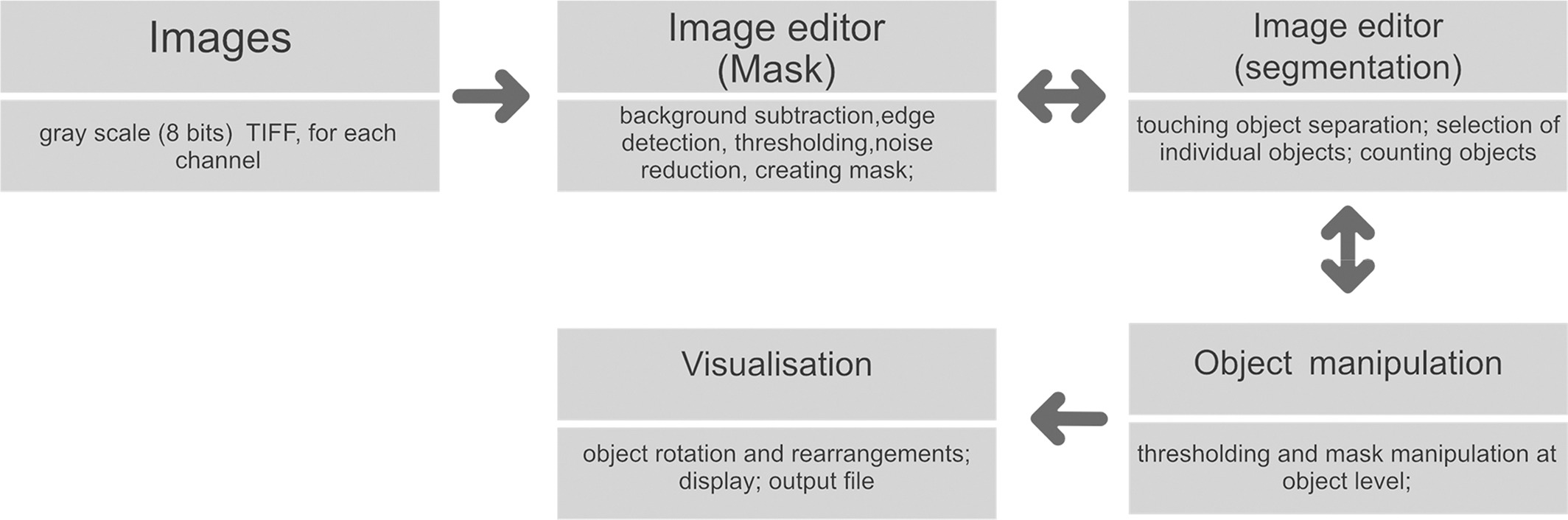
ChromaWizard is written in Python 3 and can be installed on all operating systems supporting Conda or the Python Index Wheel format. The software has no special hardware requirements and should work on any standard personal computer. ChromaWizard makes use of openCV (Open Source Computer Vision Library, version > = 3.0.0), a versatile platform which includes several libraries and hundreds of algorithms for image analysis. Additionally, NumPy[12] and scikit‐image[13] were used, which are primary platforms for scientific computing and image analysis. Finally, a GUI was generated with the Qt library version 5.7. Fig. 1 shows the overall workflow of the software.
|
ChromaWizard configuration
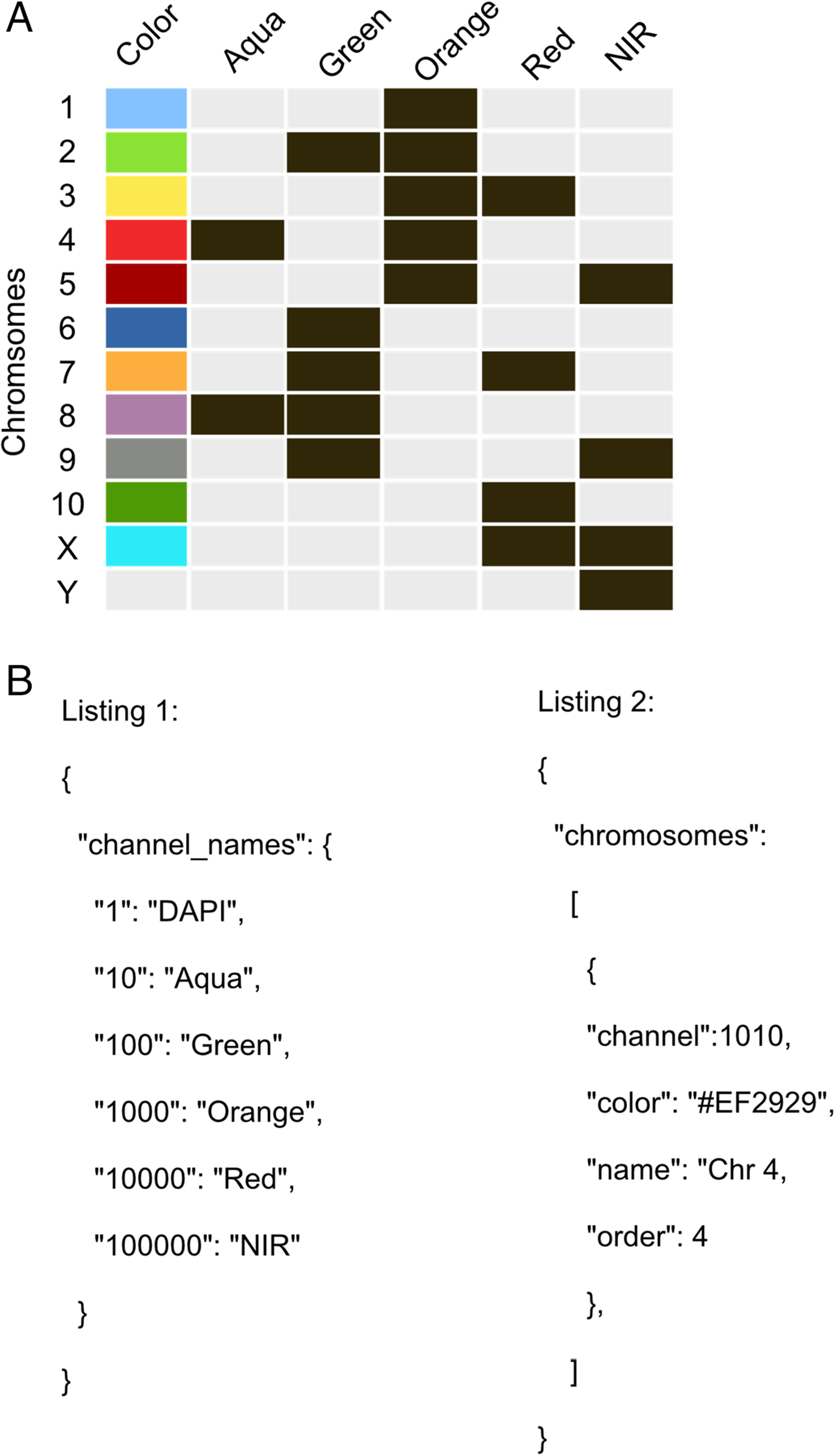
ChromaWizard is highly configurable and can be adapted to virtually any number of color channels (probes) for M‐FISH analysis in the configuration file. The default configuration file is found in the program's home folder, named “.c.default.conf.json,” after the first run. This file defines the requirements for the used labeling kit and thus for the organism of interest. The default configuration is set up for the six-channel 12XCHamster labeling kit as an example (Fig. 2A). Numbers and names of the channels and the chromosome to color association can be set in the configuration file. The configuration file is a simple text file in JSON (JavaScript Object Notation) format. A JSON editor like JSON Editor Online for changing the file is recommended; however, a standard text editor is sufficient.
|
The configuration file is divided into three parts. A “channel_names” section, a “chromosomes” section, and a “general” section for general configuration tags. The “channel_names” section configures the number, order, and names of channels used. The “chromosome” part defines all chromosomes by setting the names, colors, channel combination, and order of the chromosome in the legend view. All other configuration tags are self‐explanatory and manage general configuration tasks like setting the border thickness or border color of the selected chromosome in the separation view (Fig. 2B, above).
Image handling
The software handles grayscale (eight bits per pixel) single images in the uncompressed tagged image file format (TIFF) generated by microscopes from all major manufacturers. The prevalent image formats PNG and JPEG are also supported. Images for each channel are loaded independently, and each image must represent a specific fluorescence channel and must be loaded in the same order as defined in the config file. ChromaWizard loads image data into NumPy arrays, and objects are created for each channel, thus making original images indestructible. Furthermore, the whole project, including all images, masks, and configuration, can be saved and stored for later reanalysis or changes.
In the routinely applied chromosome painting protocols (also recommended by Metasystem), image analysis depends on 4′,6‐diamidino‐2‐phenylindole (DAPI) DNA counter-staining for efficient chromosome segmentation and generation of the global mask. In our available microscope setup, DAPI interfered with DEAC, thus limiting the use of this 12XCHamster kit. Therefore, we acquired images from all five dyes independently and one image using all fluorochromes simultaneously (to obtain a complete picture of all chromosomes, whatever their dye combination). This full karyotype image is called FULL in the following.
Thresholding and segmentations
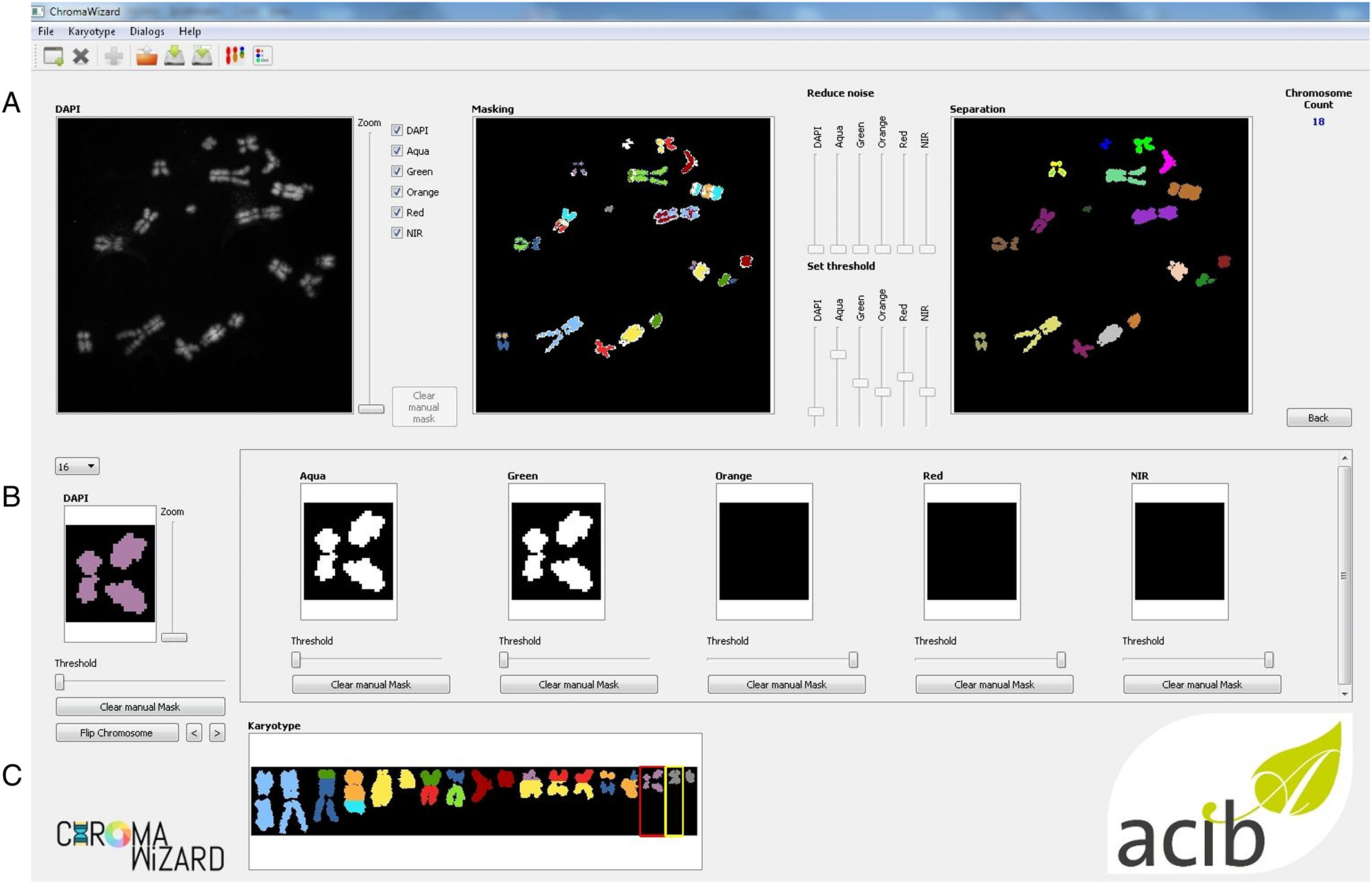
Image analysis of M‐FISH images requires pre‐processing steps such as masking and segmentation for multiplexing of the different fluorescence signals. Image thresholding and segmentations are very important aspects of multicolor analysis. ChromaWizard includes modulations so that the user can visually inspect each channel, adjust parameters for successful segmentation during each step, and control all channels independently during the entire image analysis process. Several options are offered in ChromaWizard via the GUI (Fig. 3), for example, threshold adjustment, background removal, artificial masking creation, and denoising. However, using only manual adjustments would be tedious and detract from the reproducibility of results. Thus, the program uses Otsu's algorithm[14], which automatically finds upper (foreground) and lower (background) pixel intensity thresholds by clustering‐based calculations in grayscale images with bimodal histograms, distinguishing foreground objects from background, or vice versa. The threshold set by Otsu's algorithm is a good starting value that can be used in most cases; however, as mentioned before, it can be manually refined by the user as necessary. Any signal above the threshold value is set as 1 and background as 0, converting the image into a binary image, thus enabling positive and negative mask limits. Binary masks for each channel are set similarly into a bit‐wise logically interconnected global mask. The global mask is the general FULL channel generated by counterstain, which clearly defines all elements to be analyzed and removes signals arising due to non‐specific hybridization. This is required to enhance the quality of the analysis, for example, by removal of random noise and intense background artifacts. The automated thresholding and segmentation work very well with high‐quality images; however, due to differences in imaging quality, microscopes, and/or probe quality, segmentation can fail, especially in the case of objects with saturated pixels or containing high background intensity or objects overlapping with artifacts. To handle this, the “Segmentation” image panel contains image editing tools like paint brush strokes allowing the user to remove unwanted artifacts or to divide overlapping objects after visual inspection.
|
Edge detection, filtering, counting, and visualization
ChromaWizard makes use of the global mask for automatic detection of object edges using the openCV's method connectedComponents. Overlapping and touching objects are a problem in pixel‐by‐pixel classification, which we have aimed to resolve by adding activated object editing tools in the “Segmentation” window (Fig. 3A, above). The user can select and join objects to thus combine different pieces of an object to a single entity. An example would be two chromosomes that cross each other: if the one is removed, the other remains as two separated objects that can be connected again using the software. Dependent on the total pixel size of objects and settings in the config file, small artifacts can be filtered out automatically. Finally, after separating all objects, the program counts the number of objects in the image and displays it in the right corner of the panel.
Separated objects are now displayed in the “Object” panel, where for each object the signal from each channel is displayed in the respective box (Fig. 3B, above). Objects are automatically rotated vertically, and each loaded channel is assigned to a value in a bit mask after the background correction is done in the segmentation panel. Each fluorophore is presented as a bit mask and mixed linearly according to the instructions from “color matrix” from the configuration file to assign false‐colors. ChromaWizard derives an optimal solution from the bit mask of each fluorophore after the careful noise removal at the image level. However, in practice, many factors such as non‐uniform hybridization, artifacts overlaying the object, and saturation of pixels can produce false positive signals. Thus the “Object” panel is provided with a threshold slider, where the user can adapt false positive signals at the object level, thus providing more control over the analysis. This option is particularly helpful with low-quality images for some fluorophores, which have higher cross‐talks with other channels or weak hybridization signals.
Finally, all the objects are sorted by size (from large to small) and represented for visual inspection in the “Visualization” window. Additionally, on the left side of the panel are “flip” and “move” options for user-specific presentation of chromosomes and to export the image for further analysis (Fig. 3C, above).
As an example, primary fibroblasts from Chinese hamster were analyzed from 20–30 metaphase spreads. Specificity of probes was confirmed by successful identification of 10 autosomes and X chromosomes (Fig. 4A). The apparent rearrangement of chromosome 6/9 is an artifact associated with the fact that the cell line which was used for generation of the probes had a translocation of chromosome 6–9. Thus, these regions of chromosome 6 are now labeled as chromosome 9. Similarly, we analyzed CHO‐K1 cells (Fig. 4B), where the painted karyotype clearly shows distinctive chromosomal aberrations, including both translocations and numerical changes. These changes are concurrent with previously discovered aberrations using G‐Banding and BAC hybridization reports.[15][16]
|
Summary
ChromaWizard is functional and versatile open‐source software that facilitates image processing and visualization, while also proving to be a user-friendly and robust tool. Our project aims to provide a free and lightweight program to speed up FISH and M‐FISH analysis in the absence of specialized hardware and software. It is not limited to a specific organism or a fixed karyotype. ChromaWizard is published under the General Public License (GPL 3.0) and is available at GitHub and PyPI, the Python Package Index.
Acknowledgements
The authors acknowledge support from the Austrian BMWFW, BMVIT, SFG, Standortagentur Tirol, Government of Lower Austria, and Business Agency Vienna through the Austrian FFG‐COMET‐ Funding Program.
References
- ↑ Fauth, C.; Speicher, M.R. (2001). "Classifying by colors: FISH-based genome analysis". Cytogenetics and Cell Genetics 93 (1–2): 1–10. doi:10.1159/000056937. PMID 11474168.
- ↑ Volpi, E.V.; Bridger, J.M. (2008). "FISH glossary: An overview of the fluorescence in situ hybridization technique". Biotechniques 45 (4): 385–6. doi:10.2144/000112811. PMID 18855767.
- ↑ Cui, C.; Shu, W.; Li, P. (2016). "Fluorescence in situ hybridization: Cell-based genetic diagnostic and research applications". Frontiers in Cell and Developmental Biology 4: 89. doi:10.3389/fcell.2016.00089. PMC PMC5011256. PMID 27656642. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5011256.
- ↑ Liehr, T.; Starke, H.; Weise, A. et al. (2004). "Multicolor FISH probe sets and their applications". Histology and Histopathology 19 (1): 229–37. doi:10.14670/HH-19.229. PMID 14702191.
- ↑ Schröck, E.; Veldman, T.; Padilla-Nash, H. et al. (1997). "Spectral karyotyping refines cytogenetic diagnostics of constitutional chromosomal abnormalities". Human Genetics 101 (3): 255–62. doi:10.1007/s004390050626. PMID 9439652.
- ↑ Ried, T.; Schröck, E.; Ning, Y. et al. (1998). "Chromosome painting: A useful art". Human Molecular Genetics 7 (10): 1619–26. doi:10.1093/hmg/7.10.1619. PMID 9735383.
- ↑ Liehr, T. (2018). "Basics and literature on multicolor fluorescence in situ hybridization application". fish-tl.com. http://fish-tl.com/mfish.html.
- ↑ Liehr, T.; Starke, H.; Heller, A. et al. (2006). "Multicolor fluorescence in situ hybridization (FISH) applied to FISH-banding". Cytogenetic and Genome Research 114 (3–4): 240–4. doi:10.1159/000094207. PMID 16954660.
- ↑ Schindelin, J.; Rueden, C.T.; Hiner, M.C. et al. (2015). "The ImageJ ecosystem: An open platform for biomedical image analysis". Molecular Reproduction & Development 82 (7–8): 518-529. doi:10.1002/mrd.22489. PMC PMC5428984. PMID 26153368. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5428984.
- ↑ Partipilo, G.; D'Addabbo, P.; Lacalandra, G.M. et al. (2011). "Refinement of Bos taurus sequence assembly based on BAC-FISH experiments". BMC Genomics 12: 639. doi:10.1186/1471-2164-12-639. PMC PMC3268123. PMID 22208360. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3268123.
- ↑ Vcelar, S.; Jadhav, V.; Melcher, M. et al. (2018). "Karyotype variation of CHO host cell lines over time in culture characterized by chromosome counting and chromosome painting". Biotechnology and Bioengineering 115 (1): 165-173. doi:10.1002/bit.26453. PMID 28921524.
- ↑ 12.0 12.1 van der Walt, S.; Colbert, S.C.; Varoquaux, G. (2011). "The NumPy Array: A Structure for Efficient Numerical Computation". Computing in Science & Engineering 13 (2): 22–30. doi:10.1109/MCSE.2011.37.
- ↑ van der Walt, S.; Schönberger, J.L.; Nunez-Iglesias, J. et al. (2014). "scikit-image: Image processing in Python". PeerJ 2: e453. doi:10.7717/peerj.453. PMC PMC4081273. PMID 25024921. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4081273.
- ↑ Otsu, N. (1979). "A Threshold Selection Method from Gray-Level Histograms". IEEE Transactions on Systems, Man, and Cybernetics 9 (1): 62–66. doi:10.1109/TSMC.1979.4310076.
- ↑ Deaven, L.L.; Petersen, D.F. (1973). "The chromosomes of CHO, an aneuploid Chinese hamster cell line: G-band, C-band, and autoradiographic analyses". Chromosoma 41 (2): 129–44. doi:10.1007/bf00319690. PMID 4120885.
- ↑ Cao, Y.; Kimura, S.; Itoi, T. et al. (2012). "Construction of BAC-based physical map and analysis of chromosome rearrangement in Chinese hamster ovary cell lines". Biotechnology and Bioengineering 109 (6): 1357-67. doi:10.1002/bit.24347. PMID 22012683.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation, spelling, and grammar. We also added PMCID and DOI when they were missing from the original reference.