Posted on April 24, 2018
By Shawn Douglas
Journal articles
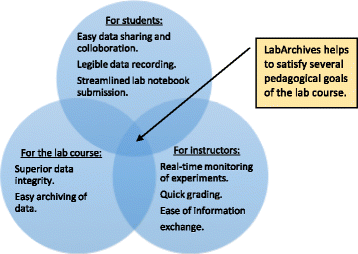
In this 2017 paper published in the
Journal of Biological Engineering, Riley
et al. of Northwestern University describe their experience with implementing the LabArchives cloud-based electronic laboratory notebook (ELN) in their bioprocess engineering laboratory course. The ultimate goal was to train students to use the ELN during the course, meanwhile promoting proper electronic record keeping practices, including good documentation practices and data integrity practices. They concluded that not only was the ELN training successful and useful but also that through the use of the ELN and its audit trail features, "a true historical record of the lab course" could be maintained so as to improve future attempts to integrate the ELN into laboratory training.
Posted on April 16, 2018
By Shawn Douglas
Journal articles
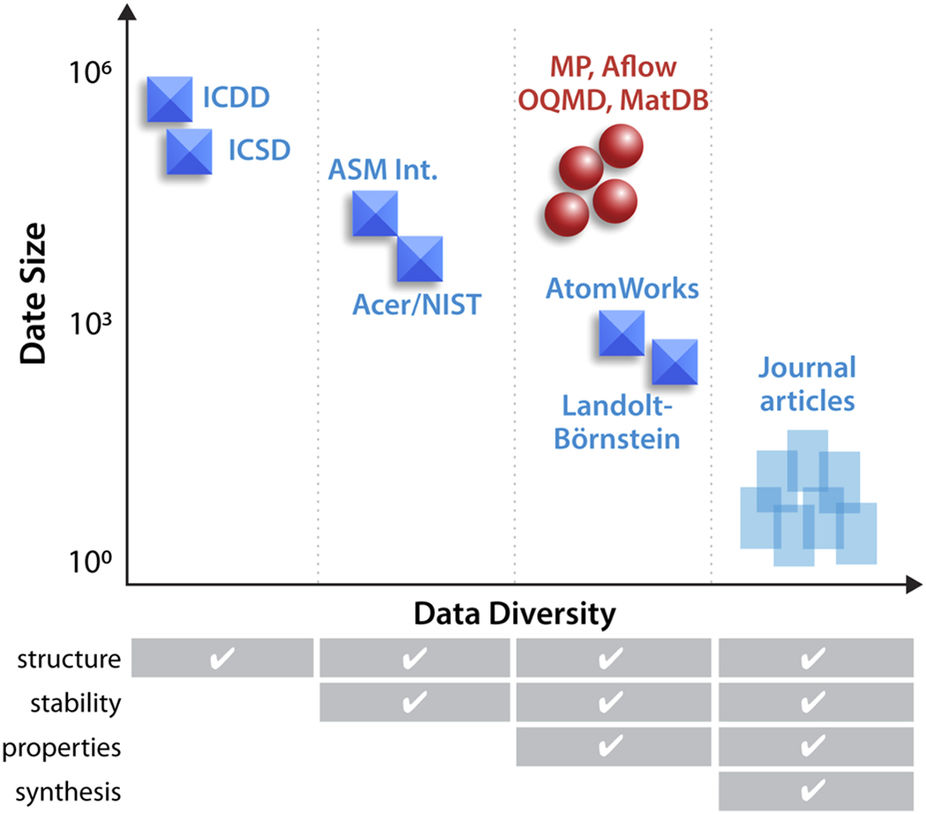
When it comes to experimental materials science, there simply aren't enough "large and diverse datasets" made publicly available say National Renewable Energy Laboratory's Zakutayev
et al. Noting this lack, the researchers built their own High Throughput Experimental Materials (HTEM) database containing 140,000 sample entries and underpinned by a custom laboratory information management system (LIMS). In this 2018 paper, the researchers discuss HTEM, the LIMS, and the how the contained sample data was derived and analyzed. They conclude that HTEM and other databases like them are "expected to play a role in emerging materials virtual laboratories or 'collaboratories' and empower the reuse of the high-throughput experimental materials data by researchers that did not generate it."
Posted on April 10, 2018
By Shawn Douglas
Journal articles
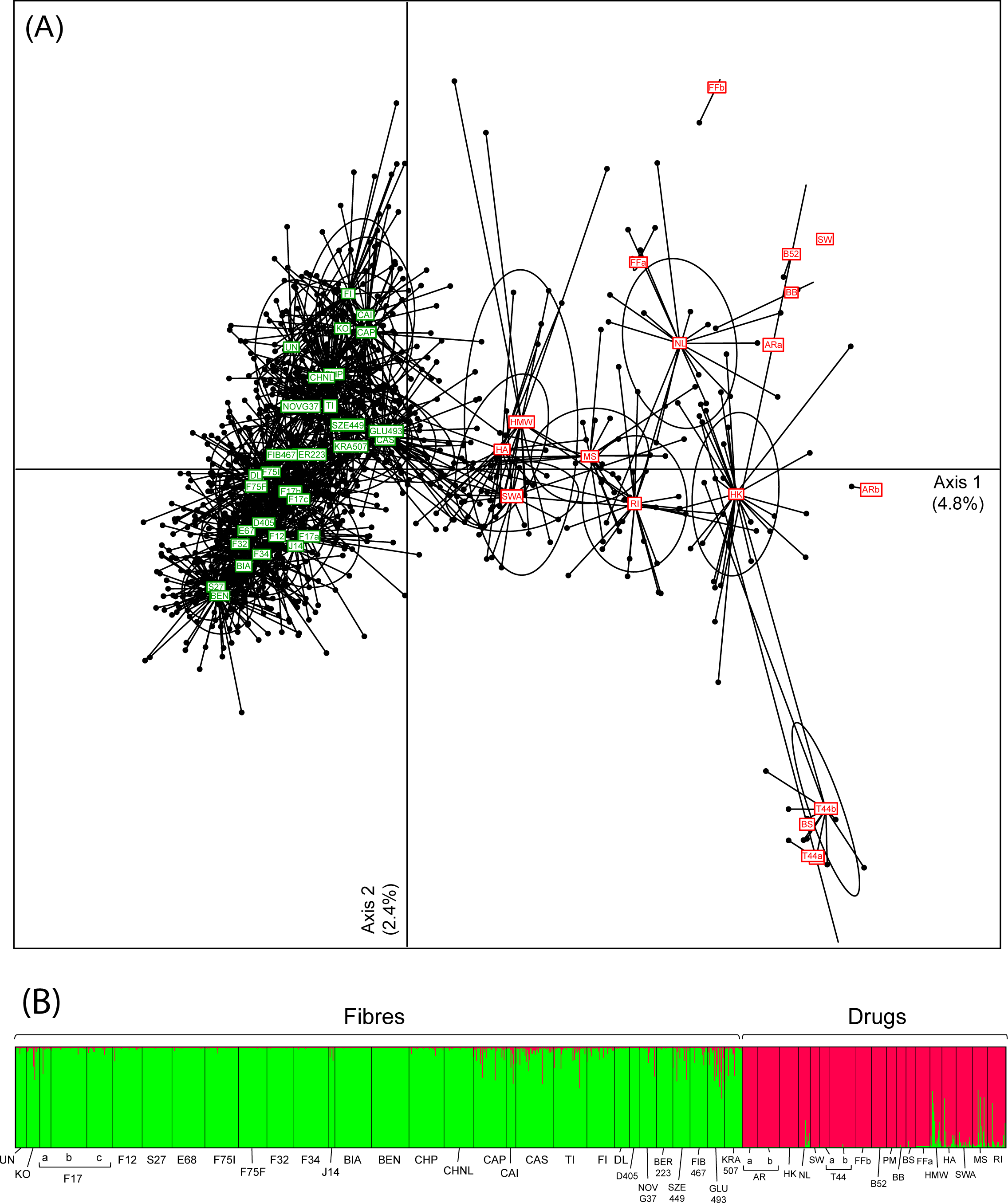
"
Cannabis ... is an iconic yet controversial crop," begin Dufresnes
et al. in this 2017 paper published in
PLOS ONE. They reveal that in actuality, due to regulations and limitations on supply, we haven't performed the same level of genetic testing on the crop in the same way we have others. Turning to next-generation sequencing (NGS) and genotyping, we can empower the field of
Cannabis forensics and other research tracks to make new discoveries. The researchers discuss their genetic database and how it was derived, ultimately concluding that databases like theirs and the "joint efforts between
Cannabis genetics experts worldwide would allow unprecedented opportunities to extend forensic advances and promote the development of the industrial and therapeutic potential of this emblematic species. "
Posted on April 3, 2018
By Shawn Douglas
Journal articles
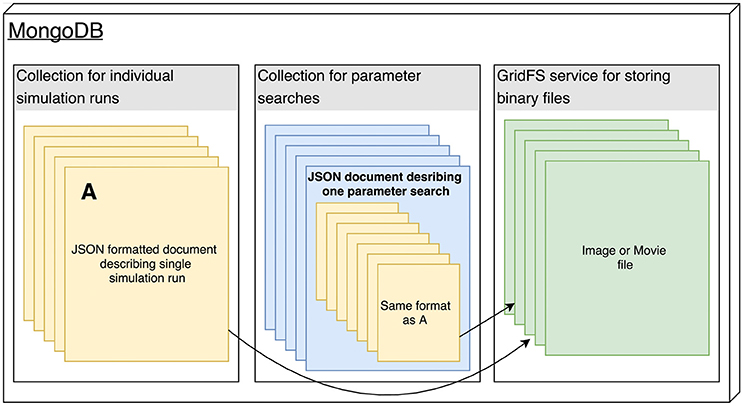
In this 2018 paper published in
Frontiers in Neuroinformatics, Antolik and Davison present Arkheia, "a web-based open science platform for computational models in systems neuroscience." The duo first describes the reasoning for creating the platform, as well as the similar systems and deficiencies. They then describe the platform architecture and its deployment, pointing out its benefits along the way. They conclude that as a whole, "Arkheia provides users with an automatic means to communicate information about not only their models but also individual simulation results and the entire experimental context in an approachable, graphical manner, thus facilitating the user's ability to collaborate in the field and outreach to a wider audience."
Posted on March 27, 2018
By Shawn Douglas
Journal articles
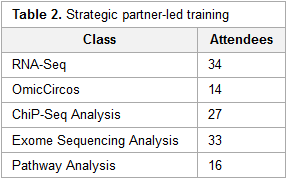
This brief case study by the National Institutes of Health's (NIH) Nathan Hosburgh takes an inside look at how the NIH took on the responsibility of bioinformatics training after the National Center for Biotechnology Information (NCBI) had to scale back its training efforts. Hosburgh provides a little background on bioinformatics and its inherent challenges. Then he delves into how the NIH—with significant help from Dr. Medha Bhagwat and Dr. Lynn Young—approached the daunting task of filling the education gap on bioinformatics, with the hope of providing "a dynamic and valuable suite of bioinformatics services to NIH and the larger medical research community well into the future."
Posted on March 20, 2018
By Shawn Douglas
Journal articles
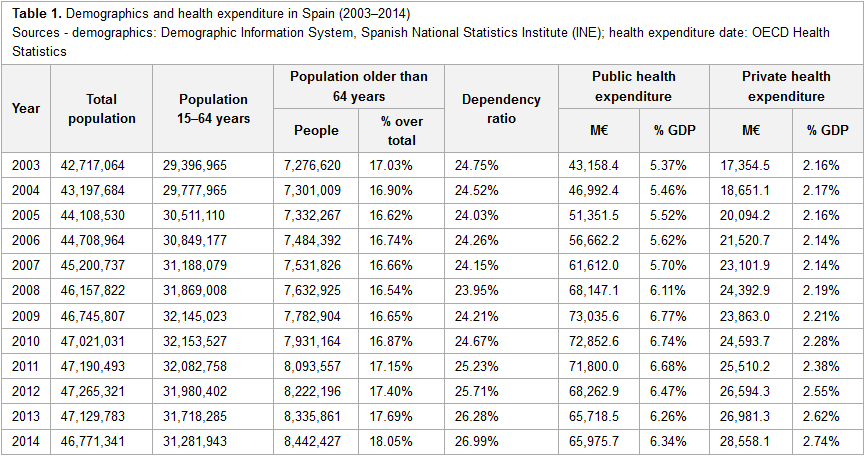
This 2018 article published in
International Journal of Interactive Multimedia and Artificial Intelligence sees Rosas and Carnicero provide their professional take—from their experience with the Spanish and other European public health system—on the benefits and challenges of implementing big data management solutions in the world of health care. After citing numbers on public and private health expenditures in relation to population, as well as reviewing literature on the subject of bid data in healthcare, the authors provide insight into some of the data systems, how they're used, and what challenges their implementation pose. They conclude that "the implementation of big data must be one of the main instruments for change in the current health system model, changing it into one with improved effectiveness and efficiency, taking into account both healthcare and economic outcomes of health services."
Posted on March 13, 2018
By Shawn Douglas
Journal articles
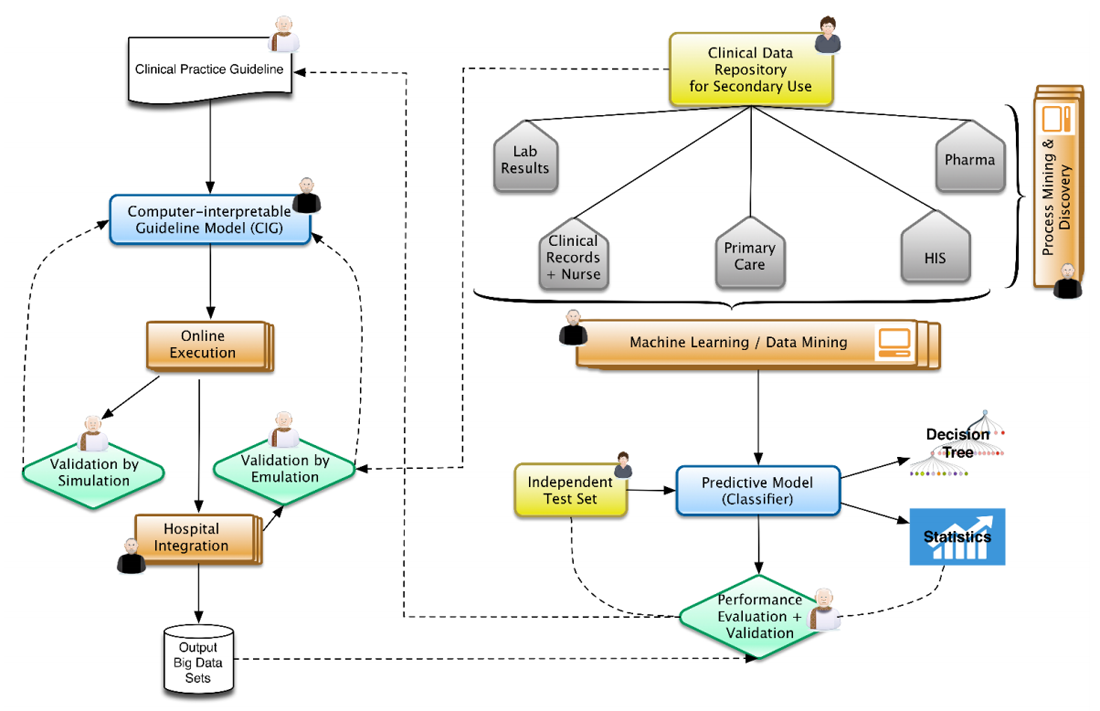
With the push for evidence-based medicine and advances in health information management over the past 30 years, the process of clinical decision making has changed significantly. However, new challenges have emerged regarding how to put the disparate data found in information management technologies such as electronic health records and clinical research databases to better use while at the same time honoring regulations and industry standards. González-Ferrer
et al. discuss these problems and how they've put solutions in place in this 2018 paper in the
International Journal of Interactive Multimedia and Artificial Intelligence. They conclude that despite the benefits of clinical decision support systems and other electronic data systems, "the development and maintenance of repositories of dissociated and normalized relevant clinical data from the daily clinical practice, the contributions of the patients themselves, and the fusion with open-access data of the social environment" will all still be required to optimize their benefits.
Posted on March 6, 2018
By Shawn Douglas
Journal articles
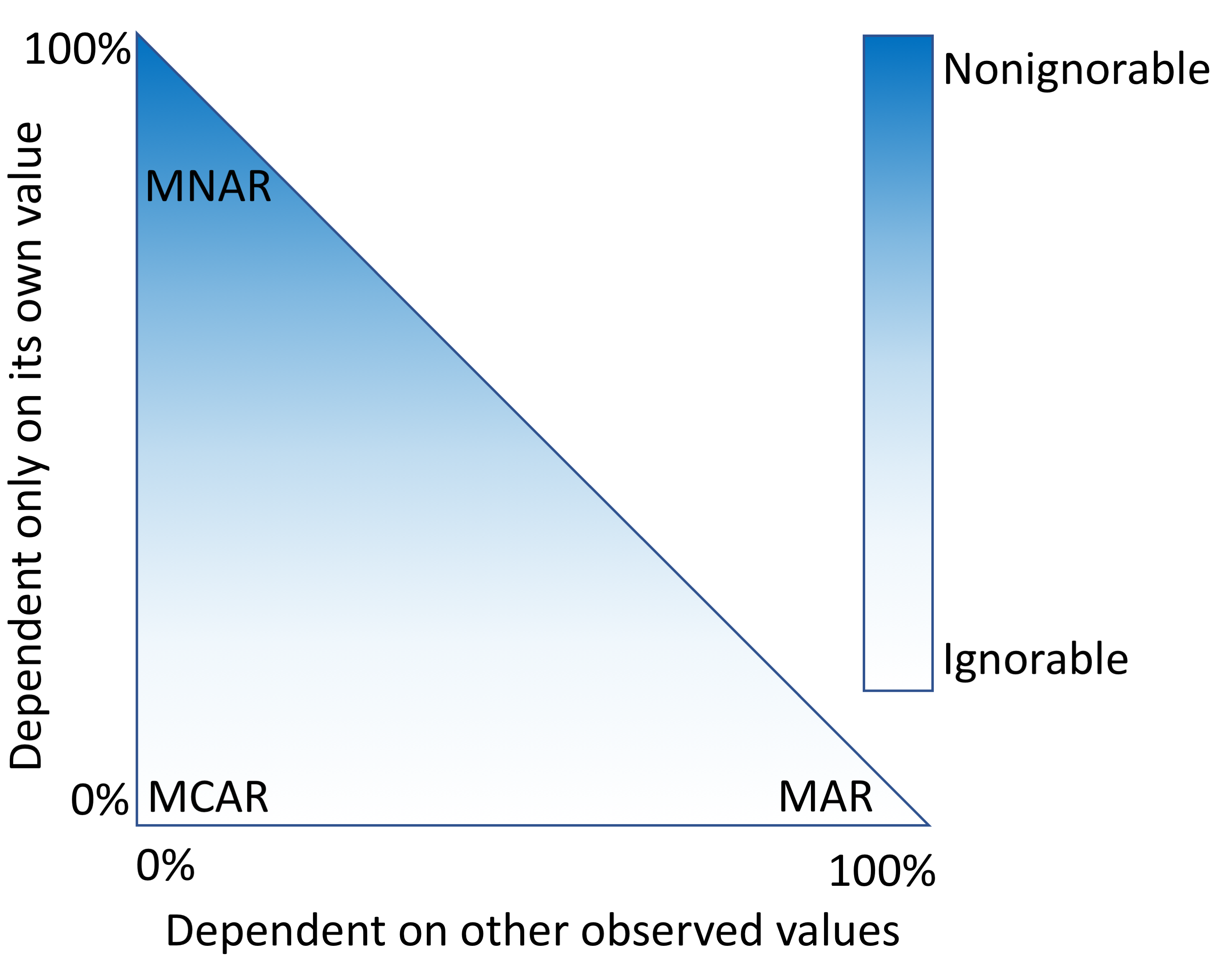
The electronic health record (EHR) was originally built with improved patient outcomes and data portability in mind, not necessarily as a resource for clinical researchers. Beaulieu-Jones
et al. point out that as a result, researchers examining EHR data often forget to take into account how missing data in the EHR records can lead to biased results. This "missingness" as they call it poses a challenge that must be corrected for. In this 2018 paper, the researchers describe a variety of correctional techniques applied to data from the Geisinger Health System's EHR, offering recommendations based on data types. They conclude that while techniques such as multiple imputation can provide "confidence intervals for the results of downstream analyses," care must be taken to assess uncertainty in any correctional technique.
Posted on February 27, 2018
By Shawn Douglas
Journal articles
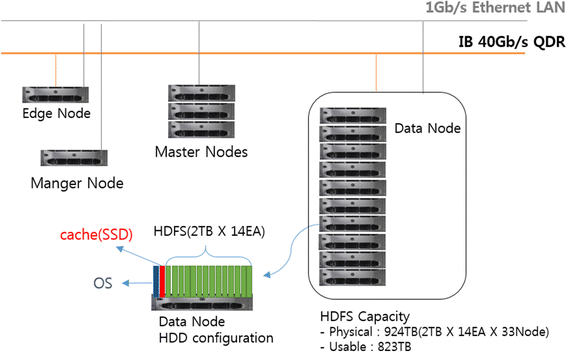
Ko
et al.of the Korean BioInformation Center discuss their Closha web service for large-scale genomic data analysis in this 2018 paper published in
BMC Bioinformatics. Noting a lack of rapid, cost-effective genomics workflow capable of running all the major genomic data analysis application in one pipeline, the researchers developed a hybrid system that can combine workflows. Additionally, they developed an add-on tool that handles the sheer size of genomic files and the speed with which they transfer, reaching transfer speeds "of up to 10 times that of normal FTP and HTTP." They conclude that "Closha allows genomic researchers without informatics or programming expertise to perform complex large-scale analysis with only a web browser."
Posted on February 20, 2018
By Shawn Douglas
Journal articles
In this 2018 paper published in
Scientific Programming, Zhu
et al. review the state of big data in the geological sciences and provide context to the challenges associated with managing that data in the cloud using China's various databases and tools as examples. Using the term "cloud-enabled geological information services" or CEGIS, they also outline the existing and new technologies that will bring together and shape how geologic data is accessed and used in the cloud. They conclude that "[w]ith the continuous development of big data technologies in addressing those challenges related to geological big data, such as the difficulties of describing and modeling geological big data with some complex characteristics, CEGIS will move towards a more mature and more intelligent direction in the future."
Posted on February 15, 2018
By Shawn Douglas
Journal articles
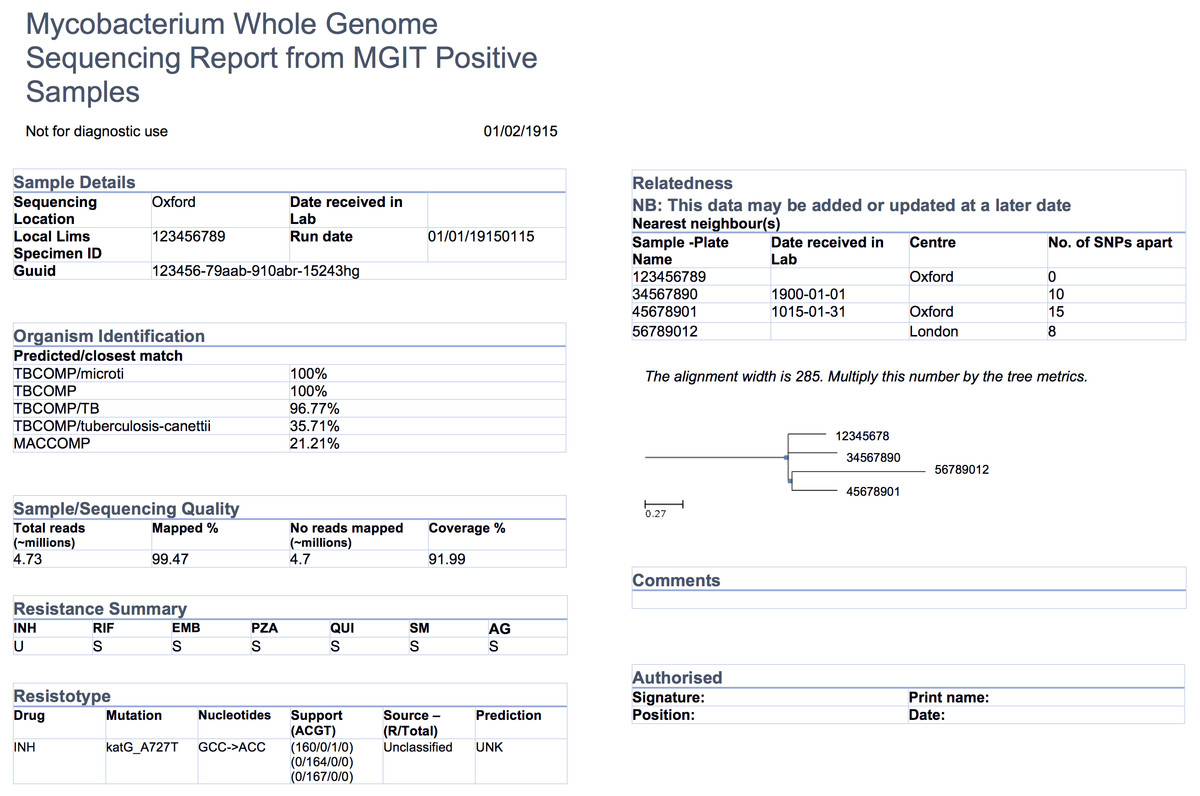
Reporting isn't as simple as casually placing key figures on a page; significant work should go into designing a report template, particularly those reporting specialized data , like that found in the world of pathogen genomics. Crisan
et al. of the University of British Columbia and the BC Centre for Disease Control looked for evidence-based guidelines on creating reports for such a specialty — specifically for tuberculosis genomic testing — and couldn't find any. So they researched and created their own. This 2018 paper details their journey towards a final report design, concluding "that the application of human-centered design methodologies allowed us to improve not only the visual aesthetics of the final report, but also its functionality, by carefully coupling stakeholder tasks, data, and constraints to techniques from information and graphic design."
Posted on February 6, 2018
By Shawn Douglas
Journal articles
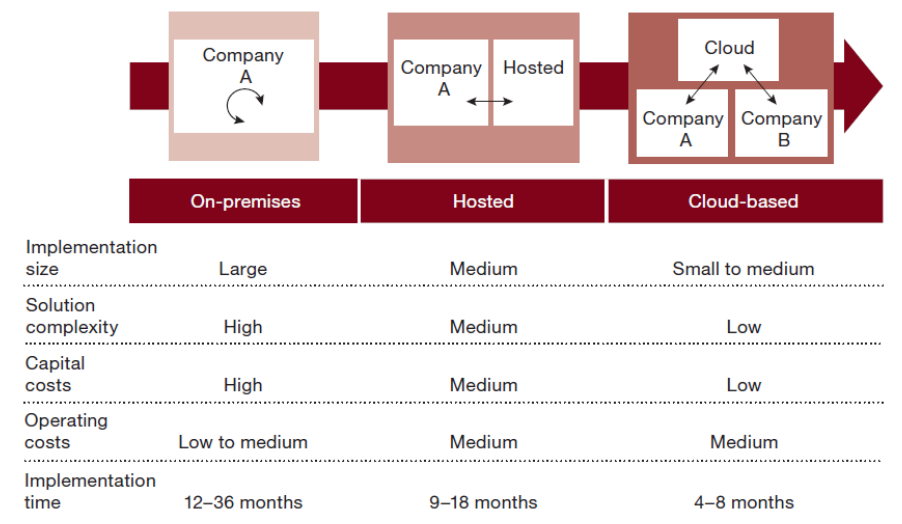
This 2017 paper by Saa
et al. examines the existing research literature concerning the status of moving an enterprise resource planning (ERP) system to the cloud. Noting both the many benefits of cloud for ERP and the data security drawbacks, the researchers discover the use of hybrid cloud-based ERPs by some organizations as a way to strike a balance, enabling them to "benefit from the agility and scalability of cloud-based ERP solutions while still keeping the security advantages from on-premise solutions for their mission-critical data."
Posted on January 30, 2018
By Shawn Douglas
Journal articles

A major goal among researchers in the age of big data is to improve data sharing and collaborative efforts. Kindler
et al. note, for example, that "specialized techniques for the production and analysis of viral metagenomes remains in a subset of labs," and other researchers likely don't know about them. As such, the authors of this 2017 paper in
F1000Research discuss the enhancement of the protocols.io online collaborative space to include more social elements "for scientists to share improvements and corrections to protocols so that others are not continuously re-discovering knowledge that scientists have not had the time or wear-with-all to publish." With its many connection points, they conclude, the software update "will allow the forum to evolve naturally given rapidly developing trends and new protocols."
Posted on January 23, 2018
By Shawn Douglas
Journal articles
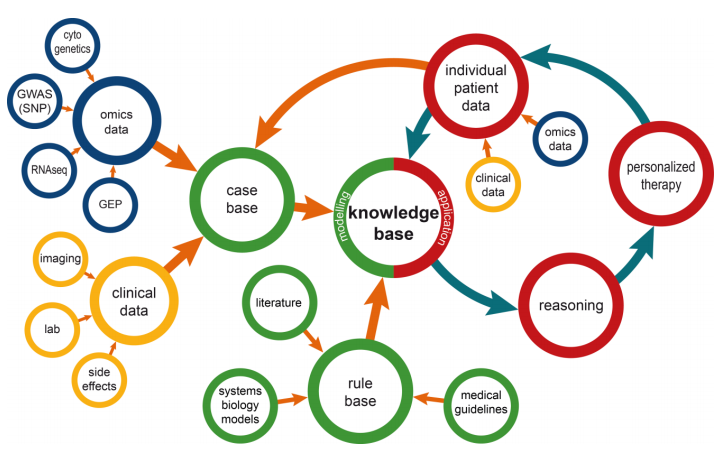
This brief paper by Ganzinger and Knaup examines the state of systems medicine, where multiple medical data streams are merged, analyzed, modeled, etc. to further how we diagnose and treat disease. They discuss the dynamic nature of disease knowledge and clinical data, as well as the problems that arise from integrating omics data into systems medicine, under the umbrella of integrating the knowledge into a usable format in tools like a clinical decision support system. They conclude that though with many benefits, "special care has to be taken to address inherent dynamics of data that are used for systems medicine; over time the number of available health records will increase and treatment approaches will change." They add that their data management model is flexible and can be used with other data management tools.
Posted on January 16, 2018
By Shawn Douglas
Journal articles
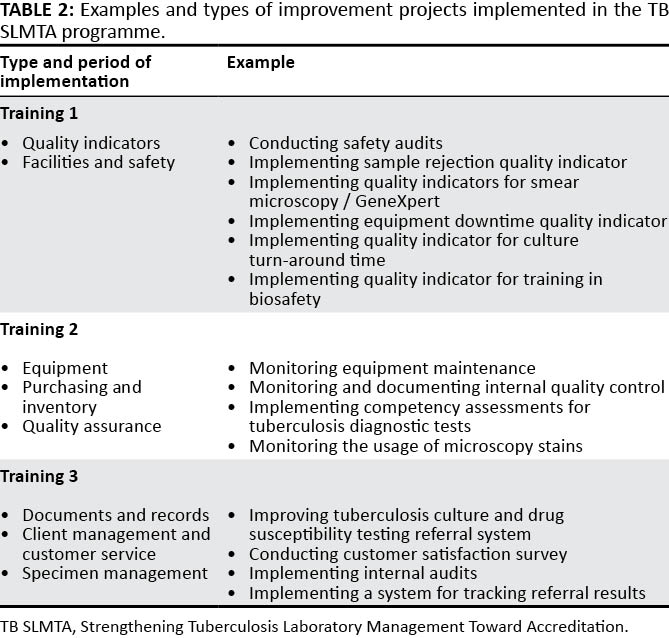
Published in early 2017, this paper by Albert
et al. discusses the development process of an accreditation program — the Strengthening Tuberculosis Laboratory Management Toward Accreditation or TB SLMTA — dedicated to better implementation of quality management systems (QMS) in tuberculosis laboratories around the world. The authors discuss the development of the curriculum, accreditation tools, and roll-out across 10 countries and 37 laboratories. They conclude that the training and mentoring program is increasingly vital for tuberculosis labs, "building a foundation toward further quality improvement toward achieving accreditation ... on the African continent and beyond."
Posted on January 8, 2018
By Shawn Douglas
Journal articles
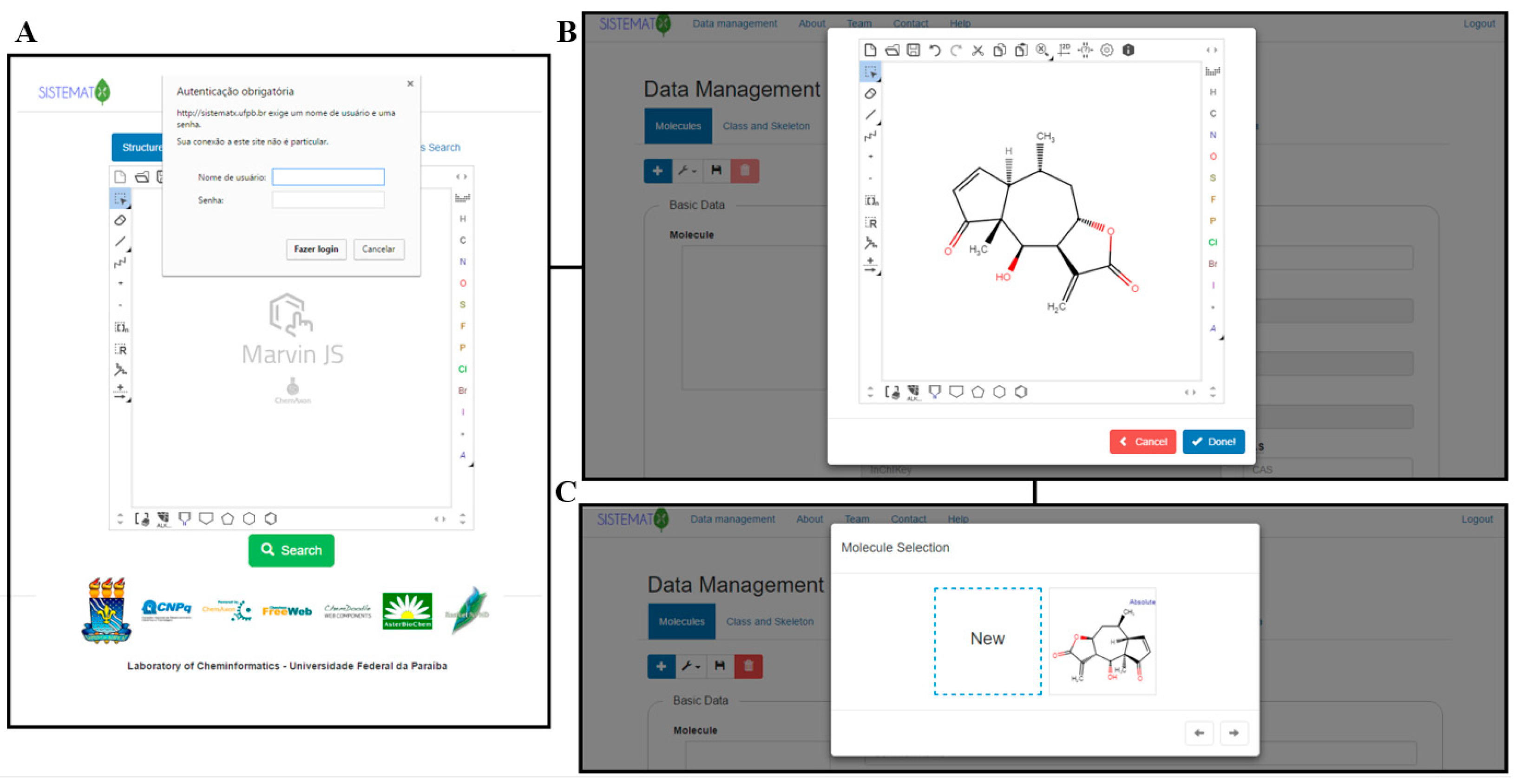
Dereplication is a chemical screening process of separation and purification that eliminates known and studied constituents, leaving other novel metabolites for future study. This is an important part of pharmaceutical and natural product development, requiring appropriate data management and retrieval for researchers around the world. However, Scotti
et al. found a lack of secondary metabolite databases that met their needs. Noting a need for complex searches, structure management, visualizations, and taxonomic rank in one package, they developed SistematX, "a modern and innovative web interface." They conclude the end result is a system that "provides a wealth of useful information for the scientific community about natural products, highlighting the location of species from which the compounds were isolated."
Posted on December 28, 2017
By Shawn Douglas
Journal articles
Social science researchers, like any other researchers, produce data in their work, both quantitative and qualitative. Unlike other sciences, social science researchers have some added difficulties in meeting the data sharing initiatives promoted by funding agencies and journals when it comes to qualitative data and how to share it "both ethically and safely." In this 2017 paper, Kirilova and Karcher describe the efforts of the Qualitative Data Repository at Syracuse University, considering the variables that went into making qualitative data available while trying to reconcile "the tension between modifying data or restricting access to them, and retaining their analytic value." They conclude that such repositories' goals should be towards "educating researchers how to be 'safe people' and how to plan for 'safe projects' – when accessing such data and using them for secondary analysis – and providing long-term 'safe settings' for the data, including via de-identification and appropriate access controls."
Posted on December 22, 2017
By Shawn Douglas
Journal articles
Metadata capture is important to scientific reproducibility argue Zehl
et al., particularly in regards to "the experiment, the data, and the applied preprocessing steps on the data." In this 2016 paper, however, the researchers demonstrate that metadata capture is not necessarily a simple task for an experimental laboratory; diverse datasets, specialized workflows, and lack of knowledge of supporting software tools all make the challenge of metadata capture more difficult. After demonstrating use cases representative of their neurophysiology lab and make a case for the open metadata Markup Language (odML), the researchers conclude that "[r]eadily available tools to support metadata management, such as odML, are a vital component in constructing [data and metadata management] workflows" in laboratories of all types.
Posted on December 12, 2017
By Shawn Douglas
Journal articles
In this 2017 paper by Plebani and Sciacovelli of the University Hospital of Padova, the duo offer their insights into the benefits and challenges of a clinical laboratory getting ISO 15189 accredited. Noting that in the European theater "major differences affect the approaches to accreditation promoted by the national bodies," the authors discuss the quality management approach that ISO 15189 prescribes and why its worth following. The conclude that while laboratories can realize "world-class quality and the need for a rigorous process of quality assurance," it still requires a high level of awareness among staff of the importance of ISO 15189 accreditation, an internal assessment plan, and well-defined, "suitable and user-friendly operating procedures."
Posted on December 4, 2017
By Shawn Douglas
Journal articles

In this 2017 paper by Curtin University's Cameron Neylon, the concepts of research data management (RDM) and research data sharing (RDS) are solidified and identified as increasingly common practices for funded research enterprises, particularly public-facing ones. But what of the implementation of these practices and the challenges associated with them? Neylon finds more than expected in his study, identifying sharp contrast among "those who saw [data management] requirements as a necessary part of raising issues among researchers and those concerned that this was leading to a compliance culture, where data management was seen as a required administrative exercise rather than an integral part of research practice." Neylon concludes with three key recommendations for researchers in general should follow to further shape how RDM and RDS are approached in research communities.
 In this 2017 paper published in the Journal of Biological Engineering, Riley et al. of Northwestern University describe their experience with implementing the LabArchives cloud-based electronic laboratory notebook (ELN) in their bioprocess engineering laboratory course. The ultimate goal was to train students to use the ELN during the course, meanwhile promoting proper electronic record keeping practices, including good documentation practices and data integrity practices. They concluded that not only was the ELN training successful and useful but also that through the use of the ELN and its audit trail features, "a true historical record of the lab course" could be maintained so as to improve future attempts to integrate the ELN into laboratory training.
In this 2017 paper published in the Journal of Biological Engineering, Riley et al. of Northwestern University describe their experience with implementing the LabArchives cloud-based electronic laboratory notebook (ELN) in their bioprocess engineering laboratory course. The ultimate goal was to train students to use the ELN during the course, meanwhile promoting proper electronic record keeping practices, including good documentation practices and data integrity practices. They concluded that not only was the ELN training successful and useful but also that through the use of the ELN and its audit trail features, "a true historical record of the lab course" could be maintained so as to improve future attempts to integrate the ELN into laboratory training.
 When it comes to experimental materials science, there simply aren't enough "large and diverse datasets" made publicly available say National Renewable Energy Laboratory's Zakutayev et al. Noting this lack, the researchers built their own High Throughput Experimental Materials (HTEM) database containing 140,000 sample entries and underpinned by a custom laboratory information management system (LIMS). In this 2018 paper, the researchers discuss HTEM, the LIMS, and the how the contained sample data was derived and analyzed. They conclude that HTEM and other databases like them are "expected to play a role in emerging materials virtual laboratories or 'collaboratories' and empower the reuse of the high-throughput experimental materials data by researchers that did not generate it."
When it comes to experimental materials science, there simply aren't enough "large and diverse datasets" made publicly available say National Renewable Energy Laboratory's Zakutayev et al. Noting this lack, the researchers built their own High Throughput Experimental Materials (HTEM) database containing 140,000 sample entries and underpinned by a custom laboratory information management system (LIMS). In this 2018 paper, the researchers discuss HTEM, the LIMS, and the how the contained sample data was derived and analyzed. They conclude that HTEM and other databases like them are "expected to play a role in emerging materials virtual laboratories or 'collaboratories' and empower the reuse of the high-throughput experimental materials data by researchers that did not generate it."
 "Cannabis ... is an iconic yet controversial crop," begin Dufresnes et al. in this 2017 paper published in PLOS ONE. They reveal that in actuality, due to regulations and limitations on supply, we haven't performed the same level of genetic testing on the crop in the same way we have others. Turning to next-generation sequencing (NGS) and genotyping, we can empower the field of Cannabis forensics and other research tracks to make new discoveries. The researchers discuss their genetic database and how it was derived, ultimately concluding that databases like theirs and the "joint efforts between Cannabis genetics experts worldwide would allow unprecedented opportunities to extend forensic advances and promote the development of the industrial and therapeutic potential of this emblematic species. "
"Cannabis ... is an iconic yet controversial crop," begin Dufresnes et al. in this 2017 paper published in PLOS ONE. They reveal that in actuality, due to regulations and limitations on supply, we haven't performed the same level of genetic testing on the crop in the same way we have others. Turning to next-generation sequencing (NGS) and genotyping, we can empower the field of Cannabis forensics and other research tracks to make new discoveries. The researchers discuss their genetic database and how it was derived, ultimately concluding that databases like theirs and the "joint efforts between Cannabis genetics experts worldwide would allow unprecedented opportunities to extend forensic advances and promote the development of the industrial and therapeutic potential of this emblematic species. "
 In this 2018 paper published in Frontiers in Neuroinformatics, Antolik and Davison present Arkheia, "a web-based open science platform for computational models in systems neuroscience." The duo first describes the reasoning for creating the platform, as well as the similar systems and deficiencies. They then describe the platform architecture and its deployment, pointing out its benefits along the way. They conclude that as a whole, "Arkheia provides users with an automatic means to communicate information about not only their models but also individual simulation results and the entire experimental context in an approachable, graphical manner, thus facilitating the user's ability to collaborate in the field and outreach to a wider audience."
In this 2018 paper published in Frontiers in Neuroinformatics, Antolik and Davison present Arkheia, "a web-based open science platform for computational models in systems neuroscience." The duo first describes the reasoning for creating the platform, as well as the similar systems and deficiencies. They then describe the platform architecture and its deployment, pointing out its benefits along the way. They conclude that as a whole, "Arkheia provides users with an automatic means to communicate information about not only their models but also individual simulation results and the entire experimental context in an approachable, graphical manner, thus facilitating the user's ability to collaborate in the field and outreach to a wider audience."
 This brief case study by the National Institutes of Health's (NIH) Nathan Hosburgh takes an inside look at how the NIH took on the responsibility of bioinformatics training after the National Center for Biotechnology Information (NCBI) had to scale back its training efforts. Hosburgh provides a little background on bioinformatics and its inherent challenges. Then he delves into how the NIH—with significant help from Dr. Medha Bhagwat and Dr. Lynn Young—approached the daunting task of filling the education gap on bioinformatics, with the hope of providing "a dynamic and valuable suite of bioinformatics services to NIH and the larger medical research community well into the future."
This brief case study by the National Institutes of Health's (NIH) Nathan Hosburgh takes an inside look at how the NIH took on the responsibility of bioinformatics training after the National Center for Biotechnology Information (NCBI) had to scale back its training efforts. Hosburgh provides a little background on bioinformatics and its inherent challenges. Then he delves into how the NIH—with significant help from Dr. Medha Bhagwat and Dr. Lynn Young—approached the daunting task of filling the education gap on bioinformatics, with the hope of providing "a dynamic and valuable suite of bioinformatics services to NIH and the larger medical research community well into the future."
 This 2018 article published in International Journal of Interactive Multimedia and Artificial Intelligence sees Rosas and Carnicero provide their professional take—from their experience with the Spanish and other European public health system—on the benefits and challenges of implementing big data management solutions in the world of health care. After citing numbers on public and private health expenditures in relation to population, as well as reviewing literature on the subject of bid data in healthcare, the authors provide insight into some of the data systems, how they're used, and what challenges their implementation pose. They conclude that "the implementation of big data must be one of the main instruments for change in the current health system model, changing it into one with improved effectiveness and efficiency, taking into account both healthcare and economic outcomes of health services."
This 2018 article published in International Journal of Interactive Multimedia and Artificial Intelligence sees Rosas and Carnicero provide their professional take—from their experience with the Spanish and other European public health system—on the benefits and challenges of implementing big data management solutions in the world of health care. After citing numbers on public and private health expenditures in relation to population, as well as reviewing literature on the subject of bid data in healthcare, the authors provide insight into some of the data systems, how they're used, and what challenges their implementation pose. They conclude that "the implementation of big data must be one of the main instruments for change in the current health system model, changing it into one with improved effectiveness and efficiency, taking into account both healthcare and economic outcomes of health services."
 With the push for evidence-based medicine and advances in health information management over the past 30 years, the process of clinical decision making has changed significantly. However, new challenges have emerged regarding how to put the disparate data found in information management technologies such as electronic health records and clinical research databases to better use while at the same time honoring regulations and industry standards. González-Ferrer et al. discuss these problems and how they've put solutions in place in this 2018 paper in the International Journal of Interactive Multimedia and Artificial Intelligence. They conclude that despite the benefits of clinical decision support systems and other electronic data systems, "the development and maintenance of repositories of dissociated and normalized relevant clinical data from the daily clinical practice, the contributions of the patients themselves, and the fusion with open-access data of the social environment" will all still be required to optimize their benefits.
With the push for evidence-based medicine and advances in health information management over the past 30 years, the process of clinical decision making has changed significantly. However, new challenges have emerged regarding how to put the disparate data found in information management technologies such as electronic health records and clinical research databases to better use while at the same time honoring regulations and industry standards. González-Ferrer et al. discuss these problems and how they've put solutions in place in this 2018 paper in the International Journal of Interactive Multimedia and Artificial Intelligence. They conclude that despite the benefits of clinical decision support systems and other electronic data systems, "the development and maintenance of repositories of dissociated and normalized relevant clinical data from the daily clinical practice, the contributions of the patients themselves, and the fusion with open-access data of the social environment" will all still be required to optimize their benefits.
 The electronic health record (EHR) was originally built with improved patient outcomes and data portability in mind, not necessarily as a resource for clinical researchers. Beaulieu-Jones et al. point out that as a result, researchers examining EHR data often forget to take into account how missing data in the EHR records can lead to biased results. This "missingness" as they call it poses a challenge that must be corrected for. In this 2018 paper, the researchers describe a variety of correctional techniques applied to data from the Geisinger Health System's EHR, offering recommendations based on data types. They conclude that while techniques such as multiple imputation can provide "confidence intervals for the results of downstream analyses," care must be taken to assess uncertainty in any correctional technique.
The electronic health record (EHR) was originally built with improved patient outcomes and data portability in mind, not necessarily as a resource for clinical researchers. Beaulieu-Jones et al. point out that as a result, researchers examining EHR data often forget to take into account how missing data in the EHR records can lead to biased results. This "missingness" as they call it poses a challenge that must be corrected for. In this 2018 paper, the researchers describe a variety of correctional techniques applied to data from the Geisinger Health System's EHR, offering recommendations based on data types. They conclude that while techniques such as multiple imputation can provide "confidence intervals for the results of downstream analyses," care must be taken to assess uncertainty in any correctional technique.
 Ko et al.of the Korean BioInformation Center discuss their Closha web service for large-scale genomic data analysis in this 2018 paper published in BMC Bioinformatics. Noting a lack of rapid, cost-effective genomics workflow capable of running all the major genomic data analysis application in one pipeline, the researchers developed a hybrid system that can combine workflows. Additionally, they developed an add-on tool that handles the sheer size of genomic files and the speed with which they transfer, reaching transfer speeds "of up to 10 times that of normal FTP and HTTP." They conclude that "Closha allows genomic researchers without informatics or programming expertise to perform complex large-scale analysis with only a web browser."
Ko et al.of the Korean BioInformation Center discuss their Closha web service for large-scale genomic data analysis in this 2018 paper published in BMC Bioinformatics. Noting a lack of rapid, cost-effective genomics workflow capable of running all the major genomic data analysis application in one pipeline, the researchers developed a hybrid system that can combine workflows. Additionally, they developed an add-on tool that handles the sheer size of genomic files and the speed with which they transfer, reaching transfer speeds "of up to 10 times that of normal FTP and HTTP." They conclude that "Closha allows genomic researchers without informatics or programming expertise to perform complex large-scale analysis with only a web browser."
 In this 2018 paper published in Scientific Programming, Zhu et al. review the state of big data in the geological sciences and provide context to the challenges associated with managing that data in the cloud using China's various databases and tools as examples. Using the term "cloud-enabled geological information services" or CEGIS, they also outline the existing and new technologies that will bring together and shape how geologic data is accessed and used in the cloud. They conclude that "[w]ith the continuous development of big data technologies in addressing those challenges related to geological big data, such as the difficulties of describing and modeling geological big data with some complex characteristics, CEGIS will move towards a more mature and more intelligent direction in the future."
In this 2018 paper published in Scientific Programming, Zhu et al. review the state of big data in the geological sciences and provide context to the challenges associated with managing that data in the cloud using China's various databases and tools as examples. Using the term "cloud-enabled geological information services" or CEGIS, they also outline the existing and new technologies that will bring together and shape how geologic data is accessed and used in the cloud. They conclude that "[w]ith the continuous development of big data technologies in addressing those challenges related to geological big data, such as the difficulties of describing and modeling geological big data with some complex characteristics, CEGIS will move towards a more mature and more intelligent direction in the future."
 Reporting isn't as simple as casually placing key figures on a page; significant work should go into designing a report template, particularly those reporting specialized data , like that found in the world of pathogen genomics. Crisan et al. of the University of British Columbia and the BC Centre for Disease Control looked for evidence-based guidelines on creating reports for such a specialty — specifically for tuberculosis genomic testing — and couldn't find any. So they researched and created their own. This 2018 paper details their journey towards a final report design, concluding "that the application of human-centered design methodologies allowed us to improve not only the visual aesthetics of the final report, but also its functionality, by carefully coupling stakeholder tasks, data, and constraints to techniques from information and graphic design."
Reporting isn't as simple as casually placing key figures on a page; significant work should go into designing a report template, particularly those reporting specialized data , like that found in the world of pathogen genomics. Crisan et al. of the University of British Columbia and the BC Centre for Disease Control looked for evidence-based guidelines on creating reports for such a specialty — specifically for tuberculosis genomic testing — and couldn't find any. So they researched and created their own. This 2018 paper details their journey towards a final report design, concluding "that the application of human-centered design methodologies allowed us to improve not only the visual aesthetics of the final report, but also its functionality, by carefully coupling stakeholder tasks, data, and constraints to techniques from information and graphic design."
 This 2017 paper by Saa et al. examines the existing research literature concerning the status of moving an enterprise resource planning (ERP) system to the cloud. Noting both the many benefits of cloud for ERP and the data security drawbacks, the researchers discover the use of hybrid cloud-based ERPs by some organizations as a way to strike a balance, enabling them to "benefit from the agility and scalability of cloud-based ERP solutions while still keeping the security advantages from on-premise solutions for their mission-critical data."
This 2017 paper by Saa et al. examines the existing research literature concerning the status of moving an enterprise resource planning (ERP) system to the cloud. Noting both the many benefits of cloud for ERP and the data security drawbacks, the researchers discover the use of hybrid cloud-based ERPs by some organizations as a way to strike a balance, enabling them to "benefit from the agility and scalability of cloud-based ERP solutions while still keeping the security advantages from on-premise solutions for their mission-critical data."
 A major goal among researchers in the age of big data is to improve data sharing and collaborative efforts. Kindler et al. note, for example, that "specialized techniques for the production and analysis of viral metagenomes remains in a subset of labs," and other researchers likely don't know about them. As such, the authors of this 2017 paper in F1000Research discuss the enhancement of the protocols.io online collaborative space to include more social elements "for scientists to share improvements and corrections to protocols so that others are not continuously re-discovering knowledge that scientists have not had the time or wear-with-all to publish." With its many connection points, they conclude, the software update "will allow the forum to evolve naturally given rapidly developing trends and new protocols."
A major goal among researchers in the age of big data is to improve data sharing and collaborative efforts. Kindler et al. note, for example, that "specialized techniques for the production and analysis of viral metagenomes remains in a subset of labs," and other researchers likely don't know about them. As such, the authors of this 2017 paper in F1000Research discuss the enhancement of the protocols.io online collaborative space to include more social elements "for scientists to share improvements and corrections to protocols so that others are not continuously re-discovering knowledge that scientists have not had the time or wear-with-all to publish." With its many connection points, they conclude, the software update "will allow the forum to evolve naturally given rapidly developing trends and new protocols."
 This brief paper by Ganzinger and Knaup examines the state of systems medicine, where multiple medical data streams are merged, analyzed, modeled, etc. to further how we diagnose and treat disease. They discuss the dynamic nature of disease knowledge and clinical data, as well as the problems that arise from integrating omics data into systems medicine, under the umbrella of integrating the knowledge into a usable format in tools like a clinical decision support system. They conclude that though with many benefits, "special care has to be taken to address inherent dynamics of data that are used for systems medicine; over time the number of available health records will increase and treatment approaches will change." They add that their data management model is flexible and can be used with other data management tools.
This brief paper by Ganzinger and Knaup examines the state of systems medicine, where multiple medical data streams are merged, analyzed, modeled, etc. to further how we diagnose and treat disease. They discuss the dynamic nature of disease knowledge and clinical data, as well as the problems that arise from integrating omics data into systems medicine, under the umbrella of integrating the knowledge into a usable format in tools like a clinical decision support system. They conclude that though with many benefits, "special care has to be taken to address inherent dynamics of data that are used for systems medicine; over time the number of available health records will increase and treatment approaches will change." They add that their data management model is flexible and can be used with other data management tools.
 Published in early 2017, this paper by Albert et al. discusses the development process of an accreditation program — the Strengthening Tuberculosis Laboratory Management Toward Accreditation or TB SLMTA — dedicated to better implementation of quality management systems (QMS) in tuberculosis laboratories around the world. The authors discuss the development of the curriculum, accreditation tools, and roll-out across 10 countries and 37 laboratories. They conclude that the training and mentoring program is increasingly vital for tuberculosis labs, "building a foundation toward further quality improvement toward achieving accreditation ... on the African continent and beyond."
Published in early 2017, this paper by Albert et al. discusses the development process of an accreditation program — the Strengthening Tuberculosis Laboratory Management Toward Accreditation or TB SLMTA — dedicated to better implementation of quality management systems (QMS) in tuberculosis laboratories around the world. The authors discuss the development of the curriculum, accreditation tools, and roll-out across 10 countries and 37 laboratories. They conclude that the training and mentoring program is increasingly vital for tuberculosis labs, "building a foundation toward further quality improvement toward achieving accreditation ... on the African continent and beyond."
 Dereplication is a chemical screening process of separation and purification that eliminates known and studied constituents, leaving other novel metabolites for future study. This is an important part of pharmaceutical and natural product development, requiring appropriate data management and retrieval for researchers around the world. However, Scotti et al. found a lack of secondary metabolite databases that met their needs. Noting a need for complex searches, structure management, visualizations, and taxonomic rank in one package, they developed SistematX, "a modern and innovative web interface." They conclude the end result is a system that "provides a wealth of useful information for the scientific community about natural products, highlighting the location of species from which the compounds were isolated."
Dereplication is a chemical screening process of separation and purification that eliminates known and studied constituents, leaving other novel metabolites for future study. This is an important part of pharmaceutical and natural product development, requiring appropriate data management and retrieval for researchers around the world. However, Scotti et al. found a lack of secondary metabolite databases that met their needs. Noting a need for complex searches, structure management, visualizations, and taxonomic rank in one package, they developed SistematX, "a modern and innovative web interface." They conclude the end result is a system that "provides a wealth of useful information for the scientific community about natural products, highlighting the location of species from which the compounds were isolated."
 Metadata capture is important to scientific reproducibility argue Zehl et al., particularly in regards to "the experiment, the data, and the applied preprocessing steps on the data." In this 2016 paper, however, the researchers demonstrate that metadata capture is not necessarily a simple task for an experimental laboratory; diverse datasets, specialized workflows, and lack of knowledge of supporting software tools all make the challenge of metadata capture more difficult. After demonstrating use cases representative of their neurophysiology lab and make a case for the open metadata Markup Language (odML), the researchers conclude that "[r]eadily available tools to support metadata management, such as odML, are a vital component in constructing [data and metadata management] workflows" in laboratories of all types.
Metadata capture is important to scientific reproducibility argue Zehl et al., particularly in regards to "the experiment, the data, and the applied preprocessing steps on the data." In this 2016 paper, however, the researchers demonstrate that metadata capture is not necessarily a simple task for an experimental laboratory; diverse datasets, specialized workflows, and lack of knowledge of supporting software tools all make the challenge of metadata capture more difficult. After demonstrating use cases representative of their neurophysiology lab and make a case for the open metadata Markup Language (odML), the researchers conclude that "[r]eadily available tools to support metadata management, such as odML, are a vital component in constructing [data and metadata management] workflows" in laboratories of all types.
 In this 2017 paper by Curtin University's Cameron Neylon, the concepts of research data management (RDM) and research data sharing (RDS) are solidified and identified as increasingly common practices for funded research enterprises, particularly public-facing ones. But what of the implementation of these practices and the challenges associated with them? Neylon finds more than expected in his study, identifying sharp contrast among "those who saw [data management] requirements as a necessary part of raising issues among researchers and those concerned that this was leading to a compliance culture, where data management was seen as a required administrative exercise rather than an integral part of research practice." Neylon concludes with three key recommendations for researchers in general should follow to further shape how RDM and RDS are approached in research communities.
In this 2017 paper by Curtin University's Cameron Neylon, the concepts of research data management (RDM) and research data sharing (RDS) are solidified and identified as increasingly common practices for funded research enterprises, particularly public-facing ones. But what of the implementation of these practices and the challenges associated with them? Neylon finds more than expected in his study, identifying sharp contrast among "those who saw [data management] requirements as a necessary part of raising issues among researchers and those concerned that this was leading to a compliance culture, where data management was seen as a required administrative exercise rather than an integral part of research practice." Neylon concludes with three key recommendations for researchers in general should follow to further shape how RDM and RDS are approached in research communities.














